library(tidyverse)
library(ggdist)
library(here)
source(here("_defaults.R"))Loading
Simulating a Galton Board
“Suppose you and a thousand of your closest friends line up in the halfway line of a soccer field.”
Ok, so the N is 1+1,000 (“you and 1000 of your closest friends”). Apparently a soccer field is 360 feet long, and an average stride length is something like 2.3 feet.
(360/2)/2.3[1] 78.26087We can get in 78 steps from the halfway line to the end of the field.
set.seed(500)
expand_grid(
person = 1:1001,
step = 1:78
) |>
mutate(
flip = sample(
c(-1, 1),
size = n(),
replace = T
)
) |>
mutate(
.by = person,
position = cumsum(flip)
) ->
galton_boardgalton_board |>
mutate(
.by = c(step, position),
n = n()
) ->
galton_boardgalton_board |>
ggplot(
aes(step, position)
)+
geom_line(
aes(group = person, color = n)
) +
scale_x_reverse()+
khroma::scale_color_bilbao(
guide = "none"
)+
coord_flip()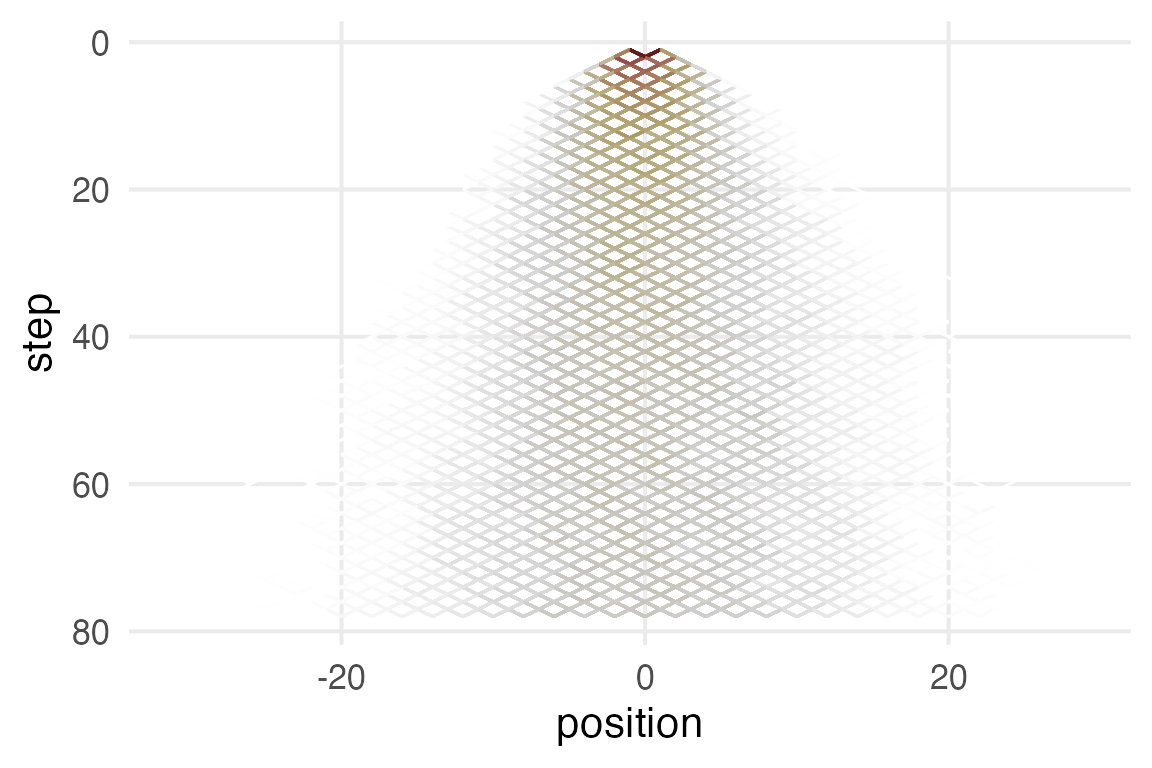
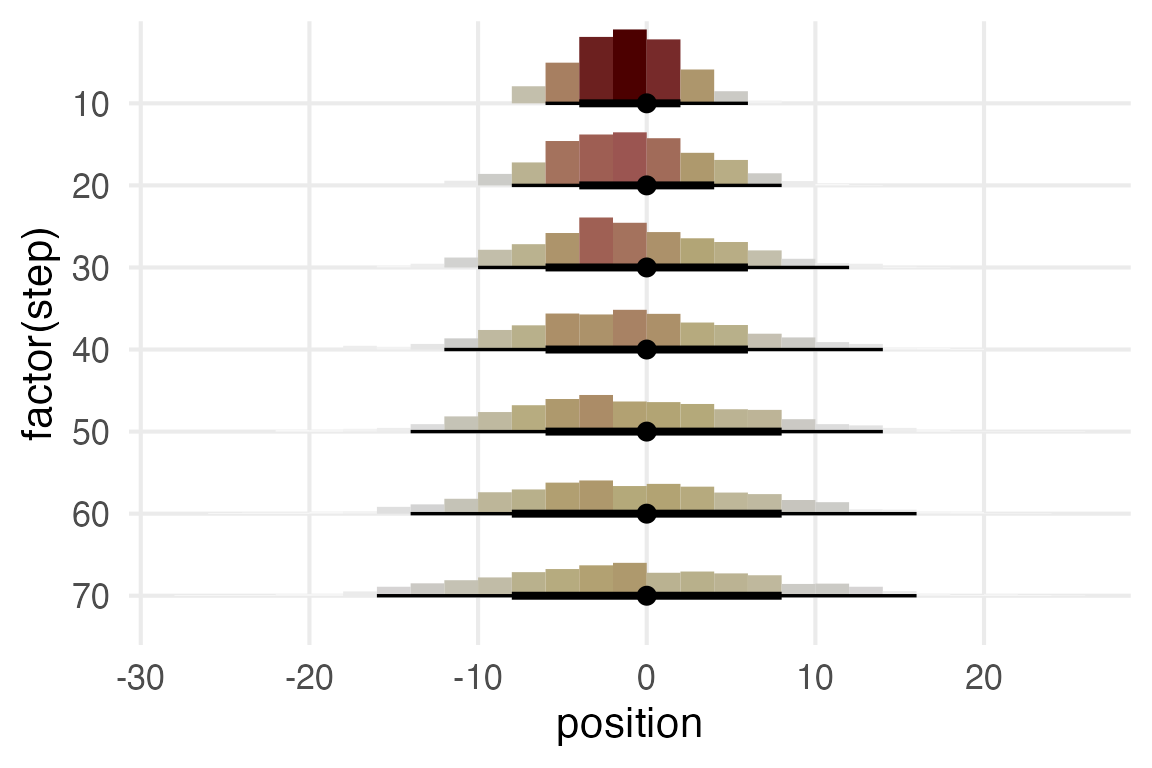
It’s hard to visualize well with the completely overlapping points. I’ll plot histograms for very 10th step.
galton_board |>
filter(step %in% seq(10, 70, by = 10)) |>
ggplot(aes(position, factor(step)))+
stat_histinterval(
breaks = breaks_fixed(width = 2),
aes(fill = after_stat(pdf))
)+
khroma::scale_fill_bilbao(
guide = "none"
)+
scale_y_discrete(
limits = factor(seq(70, 10, by = -10))
)
Infinitesimal Galton Board
Same as before, but now instead of flipping a coin for -1 and 1, values are sampled from \(\mathcal{U}(-1,1)\).
expand_grid(
person = 1:1001,
step = 1:78
) |>
mutate(
flip = runif(
n(),
-1,
1
)
) |>
mutate(
.by = person,
position = cumsum(flip)
) ->
inf_galton_boardinf_galton_board |>
ggplot(aes(step, position))+
geom_line(
aes(group = person),
alpha = 0.05
)+
scale_x_reverse()+
coord_flip()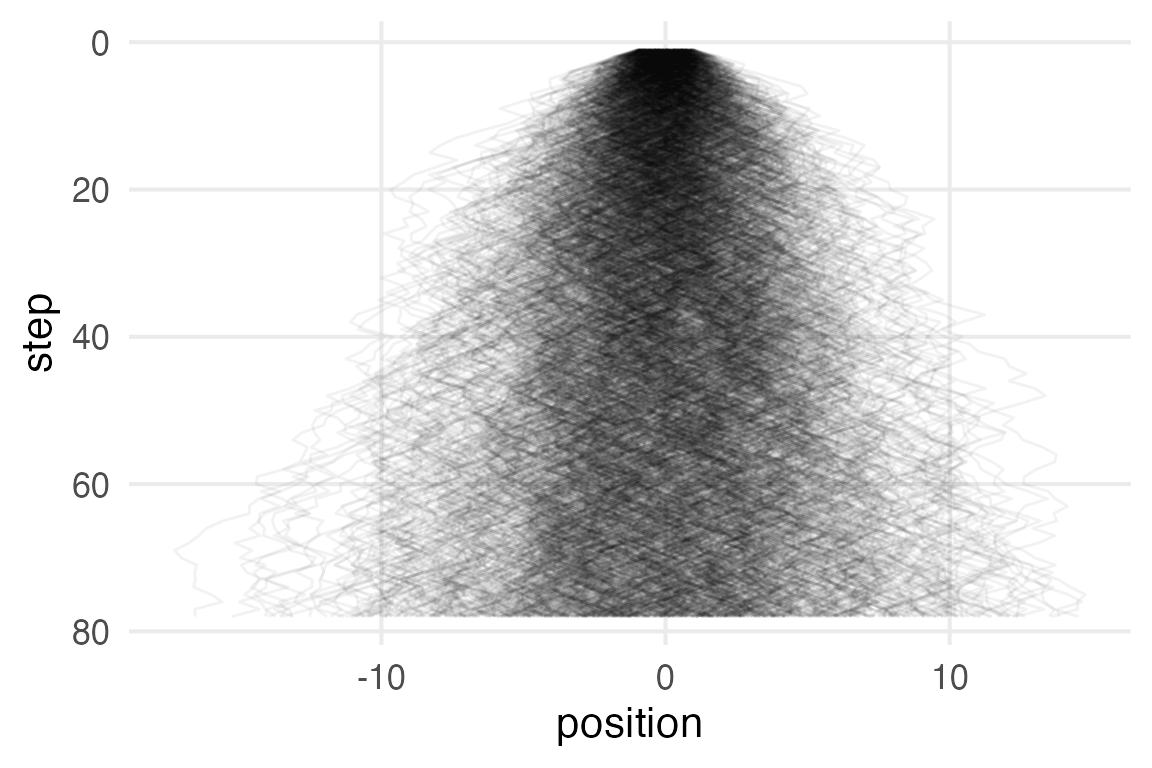
inf_galton_board |>
filter(step %in% seq(10, 70, by = 10)) |>
ggplot(aes(position, factor(step)))+
stat_slabinterval(
aes(fill = after_stat(pdf)),
fill_type = "gradient"
)+
khroma::scale_fill_bilbao(
guide = "none"
)+
scale_y_discrete(
limits = factor(seq(70, 10, by = -10))
)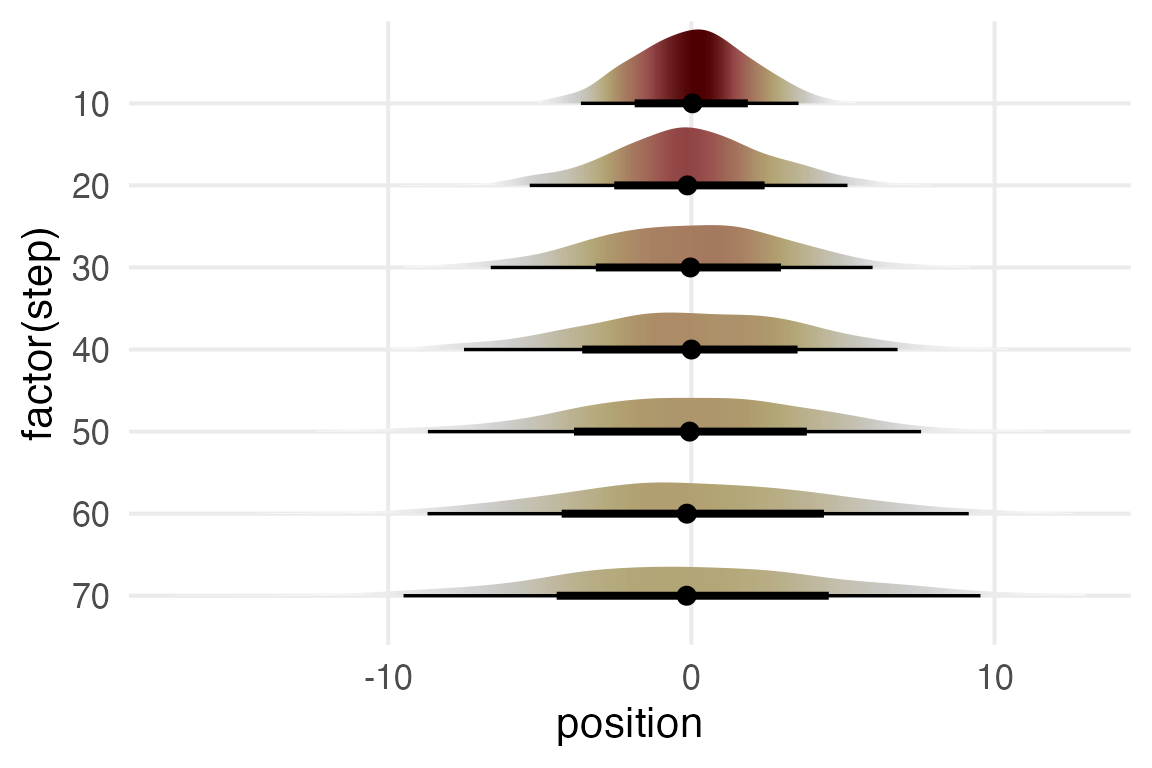
Nice.
I’m not 100% sure how to get a normal density estimate superimposed in that same plot. So I’ll fake it instead.
inf_galton_board |>
filter(step %in% seq(10, 70, by = 10)) ->
ten_steps
ten_steps |>
summarise(
.by = step,
mean = mean(position),
sd = sd(position)
) |>
nest(
.by = step
) |>
mutate(
dist = map(
data,
~tibble(
position = seq(-20, 20, length = 100),
dens = dnorm(
position,
mean = .x$mean,
sd = .x$sd
)
)
)
) |>
unnest(dist) |>
mutate(
dens_norm = dens/max(dens)
)->
distributions- 1
- Calculating the distribution parameters for each step grouping.
- 2
- Mapping over the distribution parameters to get density values in a tibble.
- 3
-
For plotting over the
stat_slab()output, normalizing the density to max out at 1.
ten_steps |>
ggplot(aes(position))+
stat_slabinterval(
aes(fill = after_stat(pdf)),
fill_type = "gradient"
)+
geom_line(
data = distributions,
aes(y = dens_norm)
)+
khroma::scale_fill_bilbao(
guide = "none"
)+
facet_wrap(
~step, labeller = label_both
)+
theme_no_y()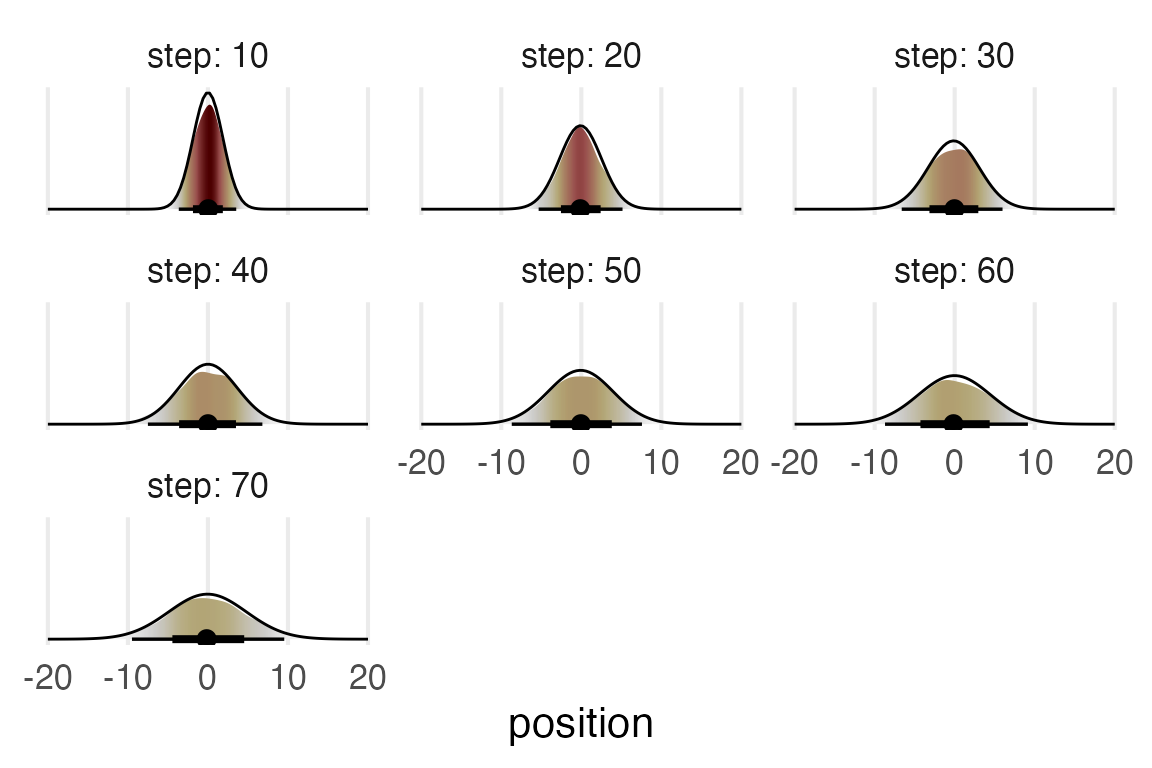
Comparing parameters
For my own interest, I wonder how much discrete sampling from -1, 1 vs the uniform distribution affects the \(\sigma\).
galton_board |>
summarise(
.by = step,
pos_sd = sd(position)
) |>
mutate(
sampling = "discrete"
) ->
galton_sd
inf_galton_board |>
summarise(
.by = step,
pos_sd = sd(position)
) |>
mutate(
sampling = "uniform"
)->
inf_galton_sdbind_rows(
galton_sd,
inf_galton_sd
) |>
ggplot(aes(step, pos_sd))+
geom_line(
aes(color = sampling),
linewidth = 1
)+
expand_limits(y = 0)+
labs(
y = expression(sigma)
)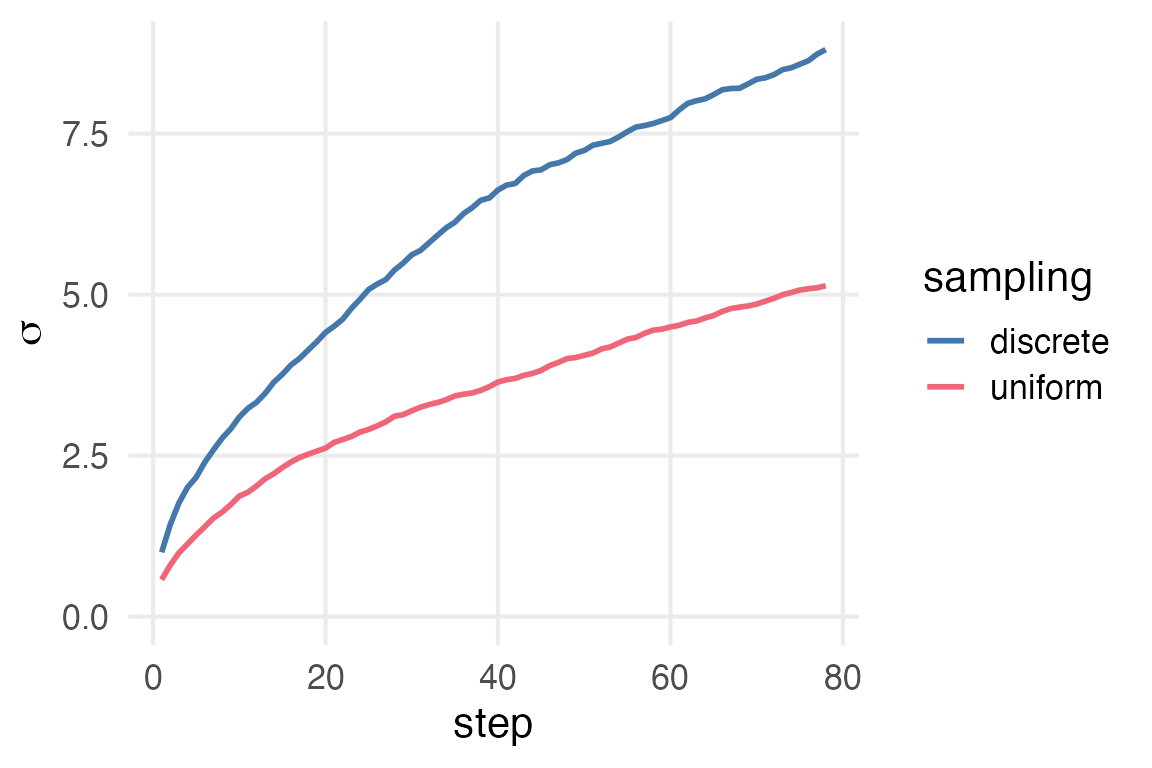
Messing around with a few obvious values of \(x\), in \(\mathcal{U}(-x,x)\), I can’t tell what would approximate the discrete sampling. 2 is too large, and 1.5 is too small. The answer is probably some horror like \(\frac{\pi}{e}\).1
Model Diagrams
Here’s the model described in the text.
\[ y_i \sim \mathcal{N}(\mu_i, \sigma) \]
\[ \mu_i = \beta x_i \]
\[ \beta \sim \mathcal{N}(0, 10) \]
\[ \sigma \sim \text{Exponential}(1) \]
He also defines a sampling distribution over \(x_1\), but idk if that’s right. Here’s my attempt at converting that into a mermaid diagram.
flowchart RL normal1["N(μᵢ, σ)"] -->|"~"| y["yᵢ"] beta["β"] --> mult1(["×"]) x[xᵢ] --> mult1 mult1 --> mu1[μᵢ] mu1 --> normal1 exp1["Exp(1)"] --"~"--> sigma1[σ] sigma1 --> normal1 normal2["N(0,10)"] --"~"--> beta
It’s ok. No quite a Kruschke diagram.
Another example.
Let me try to write out the diagram for something like y ~ x + (1|z).
\[ y \sim(\mu_i, \sigma_0) \]
\[ \mu_i = \beta_0 + \beta_1x_i + \gamma_i \]
\[ \beta_0 \sim \mathcal{N}(0,10) \]
\[ \beta_2 \sim \mathcal{N}(0,2) \]
\[ \gamma_i = \Gamma_{z_i} \]
\[ \Gamma_j \sim \mathcal{N}(0,\sigma_1) \]
\[ \sigma_0 \sim \text{Exponential}(1) \]
\[
\sigma_1 \sim \text{Exponential}(1)
\]
Geeze, idk. That double subscript feels rough, and I don’t know the convention for describing the random effects.
flowchart TD normal1["N(μᵢ, σ₀)"] --"~"--> y[yᵢ] beta0["β₀"] --> plus(["+"]) beta1["β₁"] --> plus gamma["γᵢ"] --> plus plus --> mu["μᵢ"] mu --> normal1 normal2["N(0,10)"] --"~"--> beta0 normal3["N(0,2)"] --"~"--> beta1 Gamma["Γ[zᵢ]"] --> gamma normal4["N(0, σ₁)"] --"~"--> Gamma exponent0["Exp(1)"] --"~"--> sigma0["σ₀"] sigma0 --> normal1 exponent1["Exp(1)"] --"~"--> sigma1["σ₁"] sigma1 --> normal4
Yeah, this is too tall. Will have to think about this. The Krushke style diagram is the most compressed version imo.
Footnotes
Not literally \(\frac{\pi}{e}\) though, cause that’s too small at 1.156↩︎