Code
library(tidyverse)
library(patchwork)
library(tidybayes)
library(ggdist)
library(here)
source(here("_defaults.R"))June 2, 2023
This isn’t MCMC sampling, but I’ll plot it as a line just for consistency for how MCMC chains look.
Comparing to the sampling to the original density function.
First manually
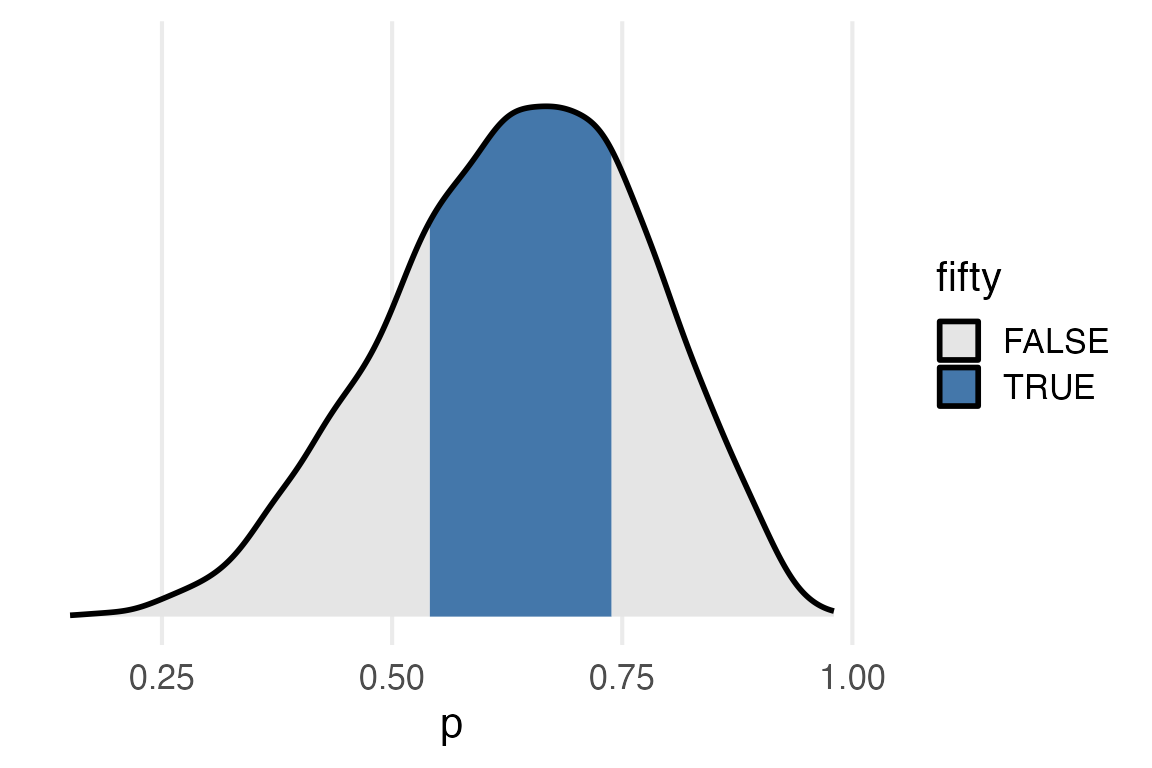
posterior_samples |>
ggplot(aes(p))+
ggdist::stat_slab(
color = "black",
aes(
fill = after_stat(
x >= fifty_quantile[1] & x <= fifty_quantile[2]
)
)
)+
labs(
fill = "fifty",
y = NULL
)+
scale_fill_manual(
values = c("grey90", ptol_blue)
)+
theme(
axis.text.y = element_blank(),
panel.grid.major.y = element_blank()
)
I think I’ll create a shortcut theme for having no y axis.
I’m not 100% sure how all of the tidybayes functions work.
# A tibble: 1 × 6
y ymin ymax .width .point .interval
<dbl> <dbl> <dbl> <dbl> <chr> <chr>
1 0.644 0.542 0.738 0.5 median qi # A tibble: 3 × 1
quantile
<dbl>
1 0.542
2 0.644
3 0.738Ok, *_qi() returns the quantile interval.
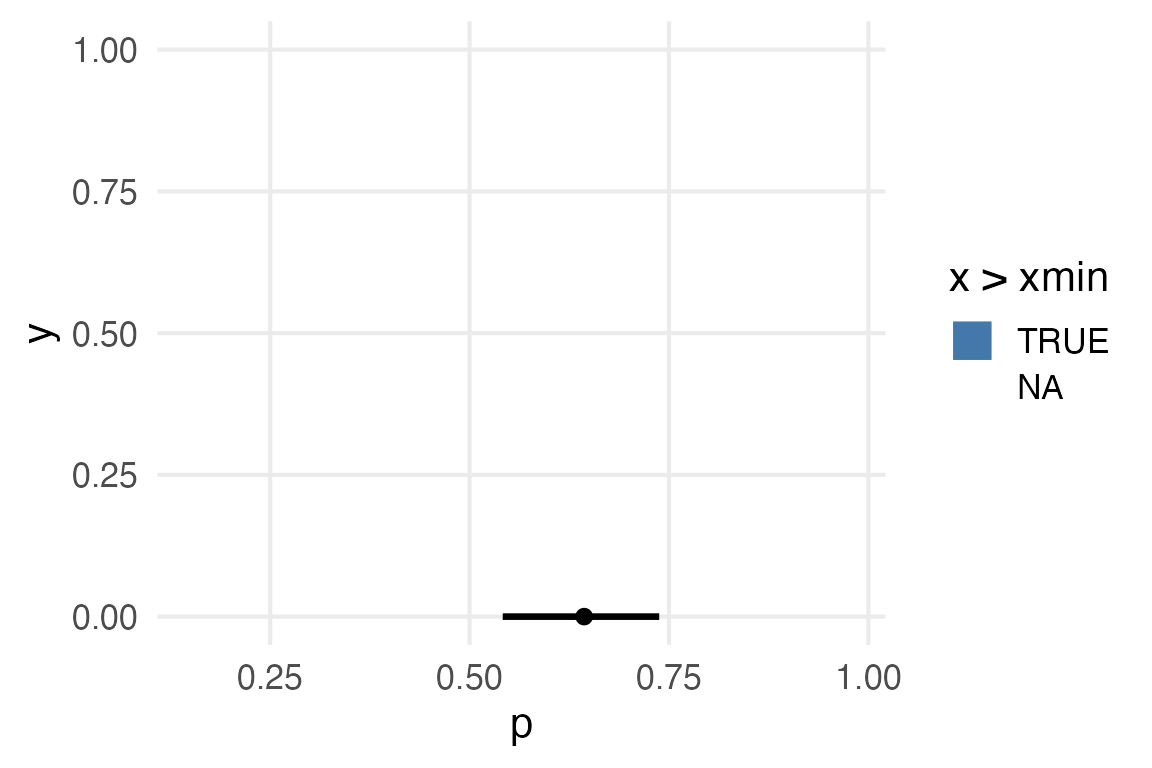
I’d like to make the plot according to the statistics calculated by stat_halfeye(), but can’t seem to get it to work.
posterior_samples |>
ggplot(aes(p))+
stat_halfeye(
.width = 0.5,
aes(
fill = after_stat(x > xmin)
)
)
I’ll just do the same filling I did before.
posterior_samples |>
ggplot(aes(p))+
stat_halfeye(
.width = 0.5,
point_interval = median_qi,
slab_color = "black",
aes(
fill = after_stat(
x >= fifty_quantile[1] & x <= fifty_quantile[2]
)
)
)+
scale_fill_manual(
values = c("grey90", ptol_blue)
)+
labs(fill = "fifty")+
theme_no_y()->
fifty_qi
fifty_qi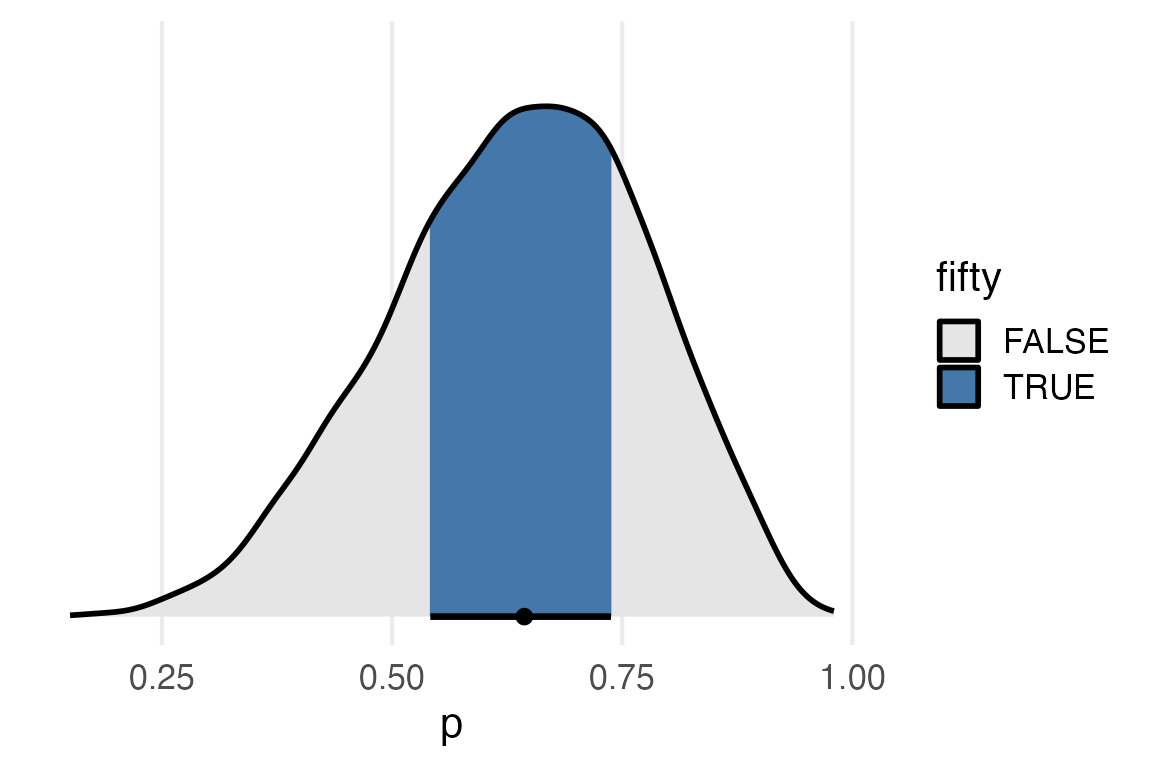
Lemme try hpdi now.
posterior_samples |>
ggplot(aes(p))+
stat_halfeye(
.width = 0.5,
point_interval = median_hdi,
slab_color = "black",
aes(
fill = after_stat(
x >= posterior_hdi$ymin &
x <= posterior_hdi$ymax
)
)
)+
scale_fill_manual(
values = c("grey90", ptol_blue)
)+
labs(fill = "fifty")+
theme_no_y()->
fifty_hdi
fifty_hdi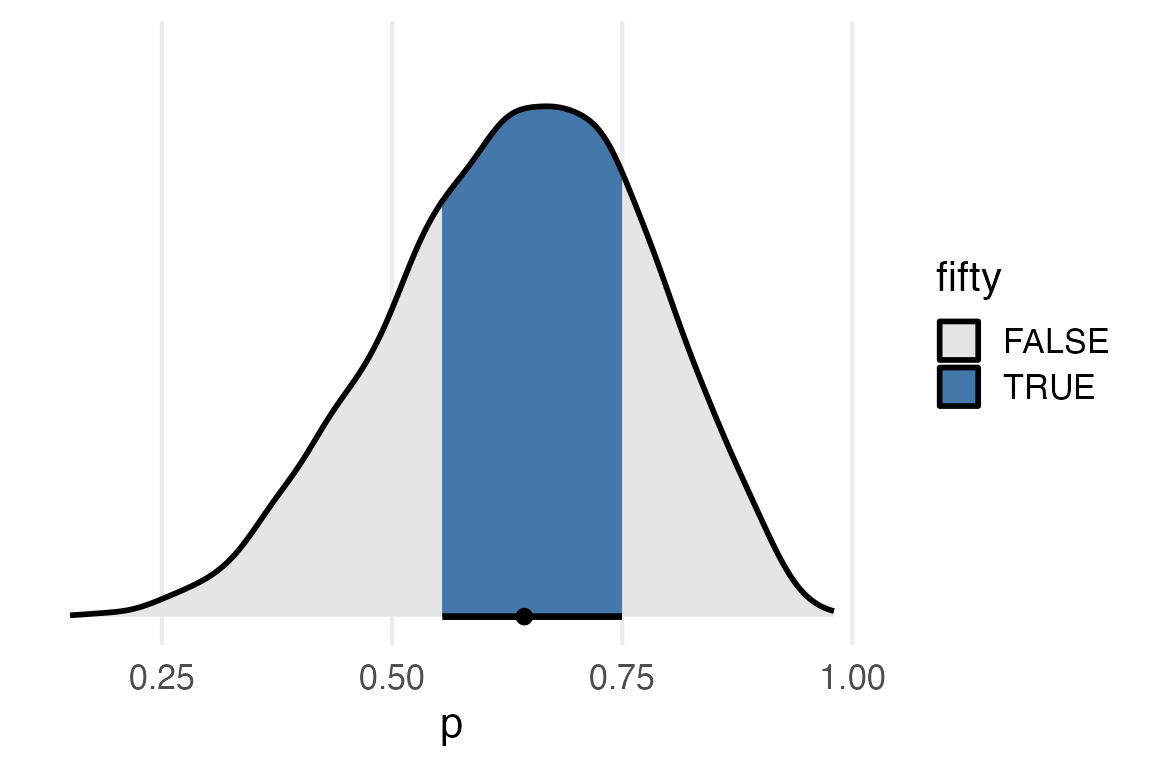
They’re very similar, but if I mix the qi fill and the hdi interval, they’re different.