library(tidyverse)
library(ggdist)
library(here)
library(ggblend)
source(here("_defaults.R"))One last post to work through different predictive distributions given 6W and 3W.
The prior predictive distribution.
The maximum likelihood.
the posterior predictive distribution.
Loading
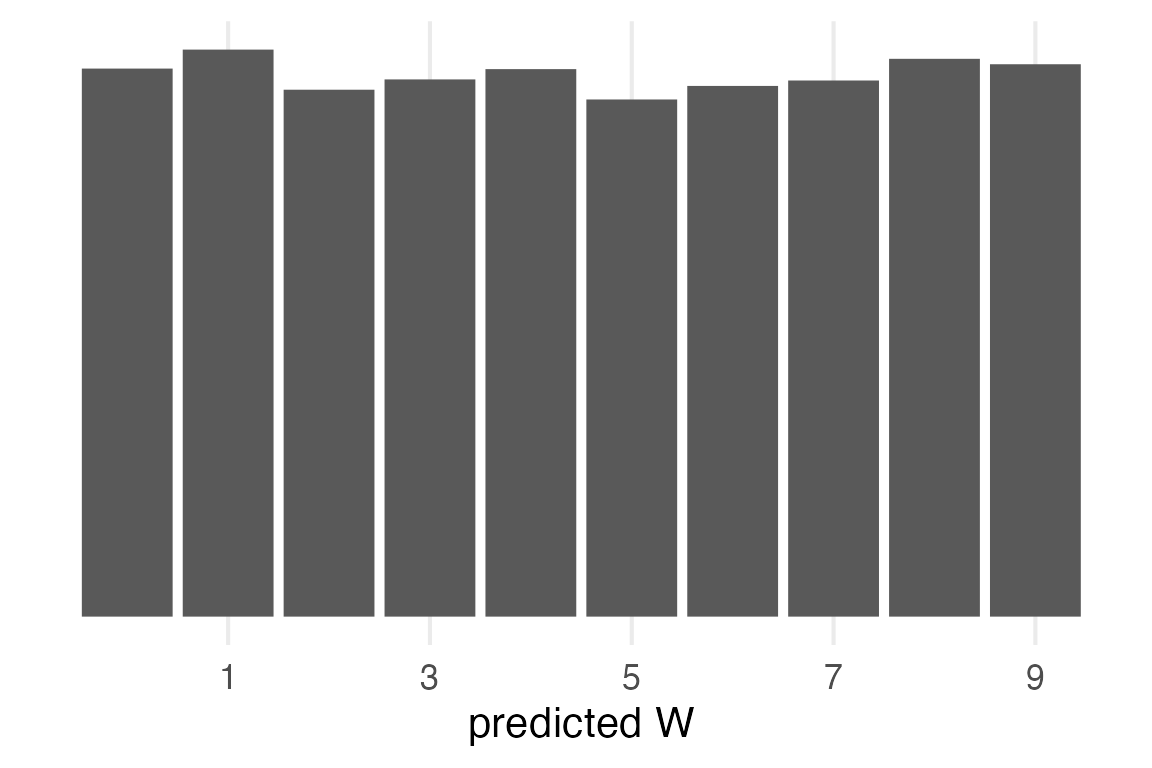
Prior predictive
The prior was a Uniform distribution between 0 and 1, or \(\mathcal{U}(0,1)\). The outcome will also be uniform, but I’ll go through all the steps for completeness.
tibble(
p = runif(1e4)
) ->
prior_sampI’ll make use of vectorization of rbinom() to generate possible samples with probabilities in prior_samp$p.
set.seed(2023)
prior_samp |>
mutate(
pred_binom = rbinom(
n(),
size = 9,
prob = p
)
) |>
count(pred_binom) |>
mutate(prob = n/sum(n)) ->
prior_pred- 1
- I’m going straight from generated samples to summarising for the distribution.
prior_pred |>
ggplot(aes(pred_binom, prob))+
geom_col()+
scale_x_continuous(
name = "predicted W",
breaks = seq(1, 9, by = 2)
)+
theme_no_y()- 1
-
I’ve moved
theme_no_y()into_defaults.Rwhich I source at the top.
Maximum Likelihood
The maximum likelihood probability of W is \(\frac{6}{9}=0.6\overline{6}\). We can get predicted values for that probability from binomial distribution.
tibble(
pred_binom = 0:9,
dens = dbinom(
pred_binom,
size = 9,
prob = 6/9
),
prob = dens
) ->
ml_pred
ml_pred |>
ggplot(aes(pred_binom, prob))+
geom_col()+
scale_x_continuous(
name = "predicted W",
breaks = seq(1, 9, by = 2)
)+
theme_no_y()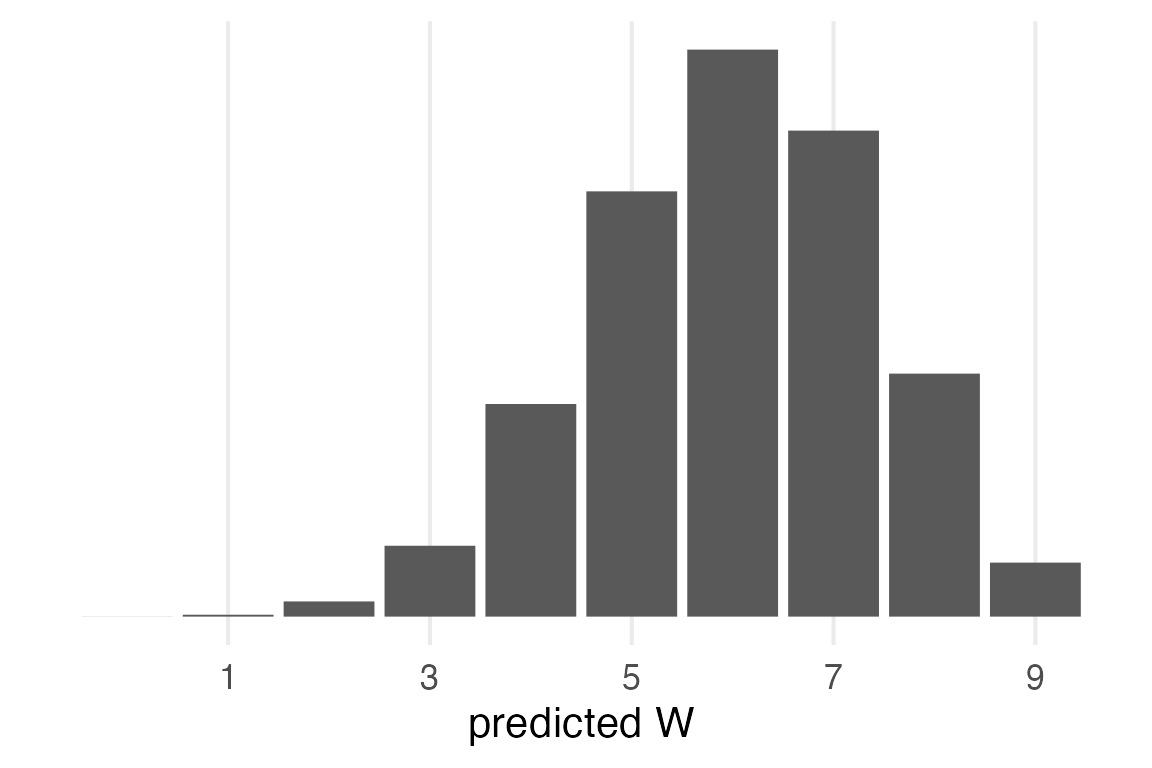
Posterior Prediction
For the posterior prediction, first we sample probabilities from the beta distribution
\[ p \sim \text{Beta}(W+1, L+1) \]
(a.k.a. dbeta())
Then for each \(p\), we generate predictions from the binomial distribution.
\[ Y|p \sim \text{Bin}(9,p) \]
This chapter has been doing grid sampling, but I’ll just use the rbeta() function for simplicity.
tibble(
p = rbeta(1e4, 6+1, 3+1)
) ->
posterior_sampThen, it’s the same operation as the prior predictive distribution.
posterior_samp |>
mutate(
pred_binom = rbinom(
n(),
size = 9,
prob = p
)
) |>
count(pred_binom) |>
mutate(prob = n/sum(n)) ->
posterior_predposterior_pred |>
ggplot(aes(pred_binom, prob))+
geom_col() +
scale_x_continuous(
name = "predicted W",
breaks = seq(1, 9, by = 2)
) +
theme_no_y()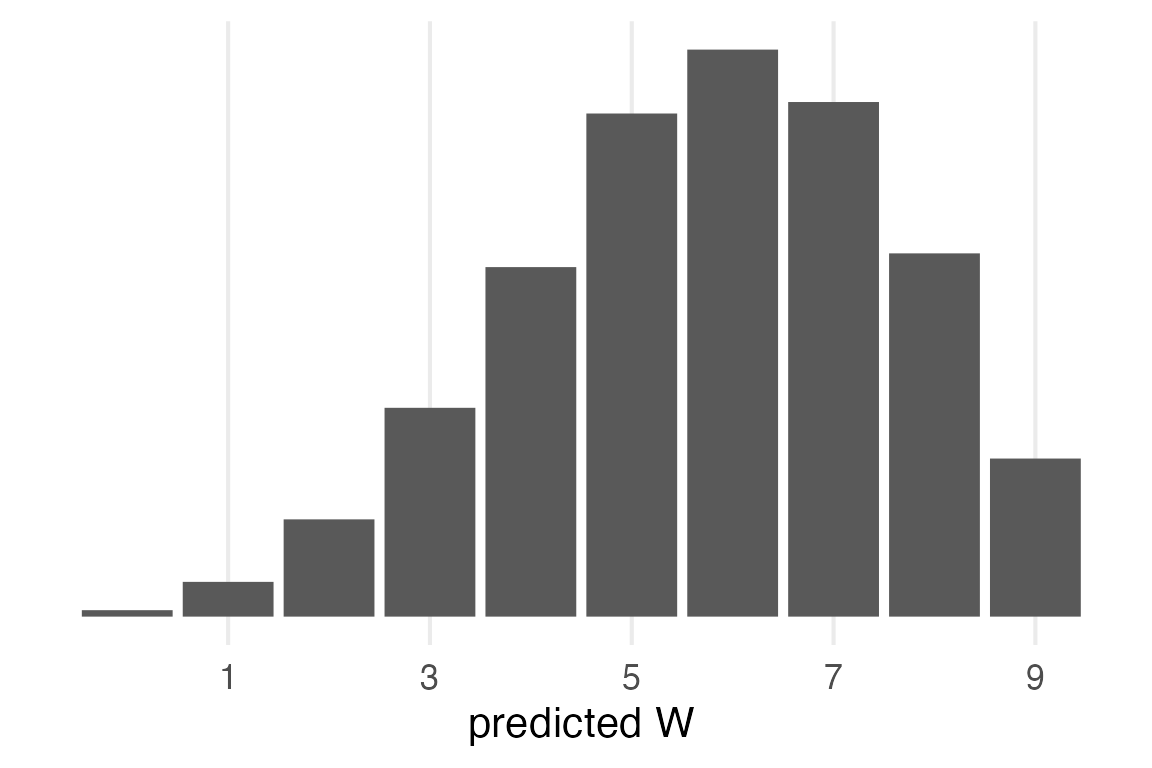
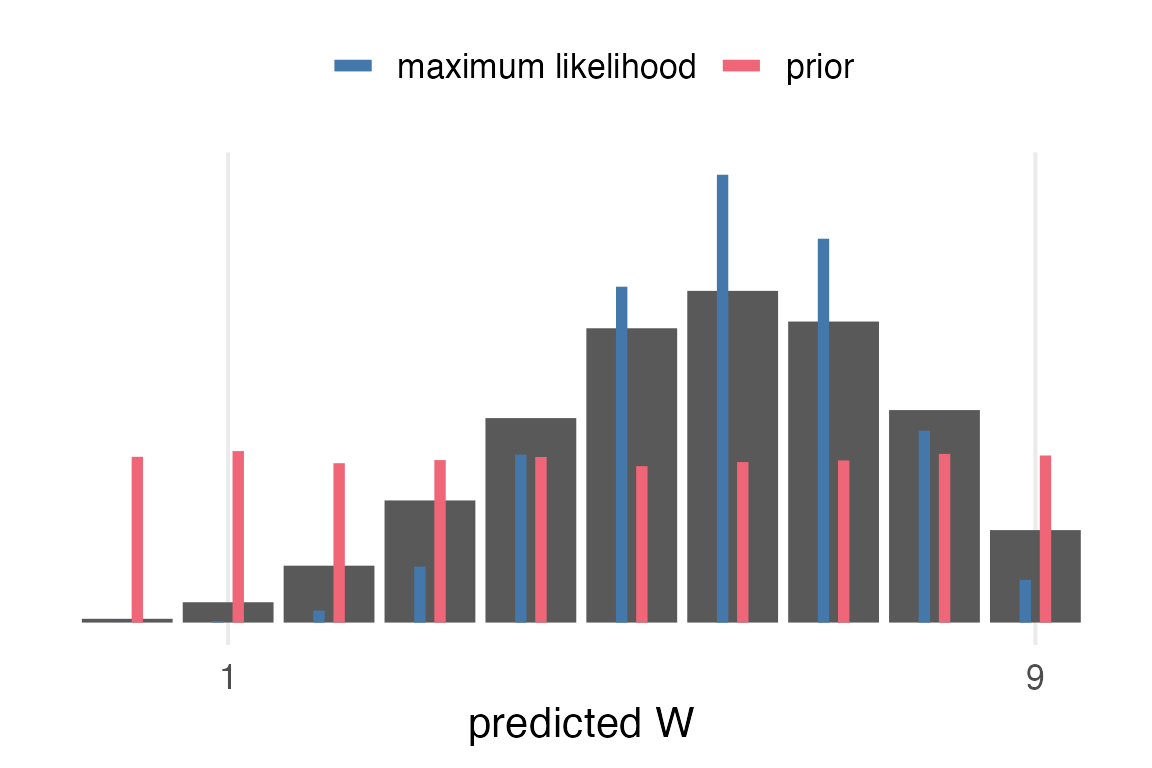
Comparisons
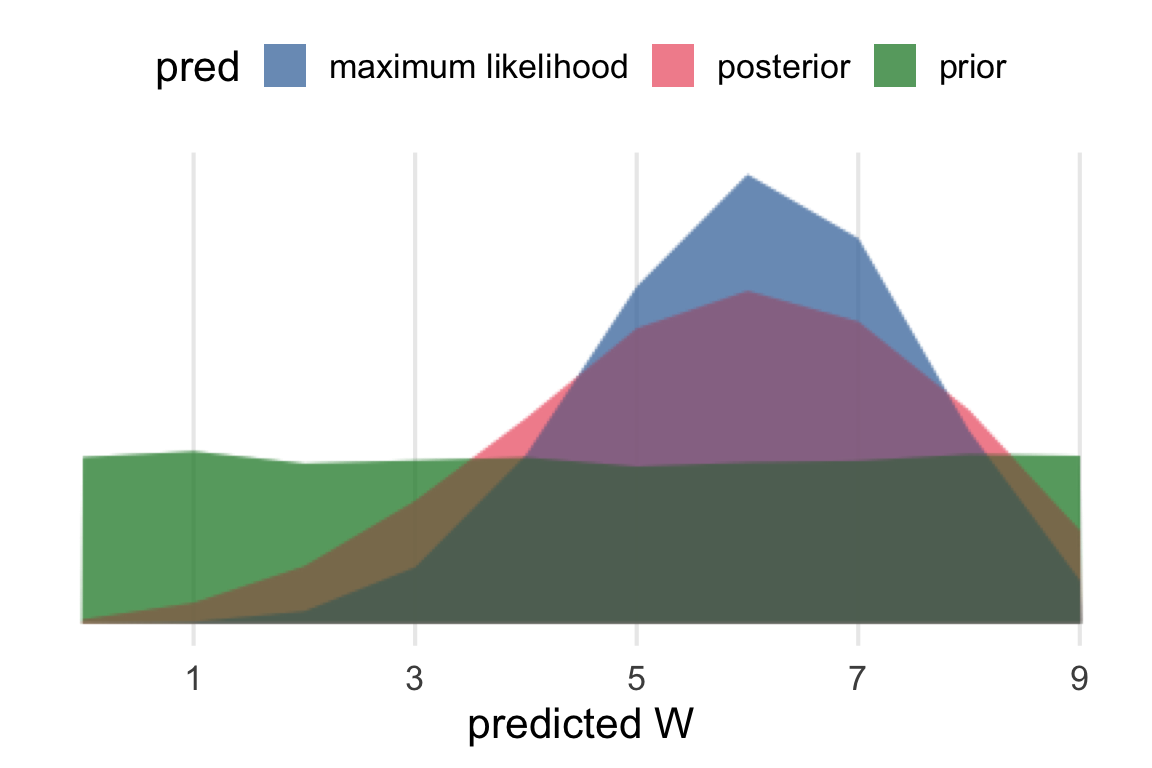
Now, I want to compare the posterior predictive distribution to the maximum likelihood and the prior. This is one concept I had, which has the prior and the ML values as thinner bars within the posterior predictive bars.
posterior_pred |>
ggplot(aes(pred_binom, prob))+
geom_col()+
geom_segment(
data = ml_pred,
aes(
x = pred_binom-0.1,
xend = pred_binom-0.1,
yend = 0,
color = "maximum likelihood"
),
linewidth = 2
)+
geom_segment(
data = prior_pred,
aes(
x = pred_binom+0.1,
xend = pred_binom+0.1,
yend = 0,
color = "prior"
),
linewidth = 2
)+
scale_x_continuous(
breaks = seq(1, 9, length = 2)
)+
labs(
color = NULL,
x = "predicted W"
)+
theme_no_y()+
theme(
legend.position = "top"
)
Overplotting them is also a possibility, and a good occasion to test out {ggblend}. I had some issues getting this to work with the available graphic devices & quarto.
```{r}
#| dev: "png"
#| dev-args:
#| - type: "cairo"
bind_rows(
posterior_pred |>
mutate(pred = "posterior"),
ml_pred |>
mutate(pred = "maximum likelihood"),
prior_pred |>
mutate(pred = "prior")
) |>
ggplot(aes(pred_binom, prob, fill = pred))+
geom_area(position = "identity", alpha = 0.5) *
(blend("lighten") + blend("darken")) +
scale_x_continuous(
name = "predicted W",
breaks = seq(1, 9, by = 2)
)+
theme_no_y()+
theme(
legend.position = "top"
)
```
Visualizing the posterior prediction process
posterior_samp |>
slice(1:4) |>
mutate(
pred_binom = rbinom(
n(),
size = 9,
prob = p
)
) |>
arrange(
p
) ->
example_sampexample_samp |>
mutate(
dens = dbeta(p, 6+1, 3+1),
n = row_number()
)->
example_samp
tibble(
p = seq(0, 1, length = 100),
dens = dbeta(p, 6+1, 3+1)
) |>
ggplot(aes(p, dens))+
geom_area(fill = "grey") +
geom_segment(
data = example_samp,
aes(
xend = p,
yend = 0,
color = factor(n)
),
linewidth = 1.5,
lineend = "round",
show.legend = F
)+
theme_no_y()+
labs(
title = "Posterior"
)->
posterior_plotThis involved messing around with {rlang} data masking that I still don’t quite follow. Both the p and pred arguments had to be (just once!) prefixed with !!, but not n.
sample_plot_fun <- function(p, pred, n){
this_scale <- as.vector(khroma::color("bright")(4))
tibble(
ws = 0:9,
dens = dbinom(ws, size = 9, prob = !!p)
) |>
ggplot(aes(ws, dens))+
geom_col(
aes(fill = ws == !!pred)
)+
scale_fill_manual(
values = c("grey", this_scale[n]),
guide = "none"
) +
scale_x_continuous(
breaks = seq(1,9, by = 2)
)+
labs(
x = "prediction",
title = str_glue("p = {round(p, digits = 2)}")
) +
theme_no_y()+
theme(
plot.title = element_text(size = 10)
)
}example_samp |>
rowwise() |>
mutate(
plot = list(
sample_plot_fun(p, pred_binom, n)
)
)->
samp_plotslibrary(patchwork)Going to use some patchwork and purr::reduce() fanciness.
posterior_plot/
(samp_plots |>
ungroup() |>
pull(plot) |>
reduce(.f = `+`) +
plot_layout(nrow = 1))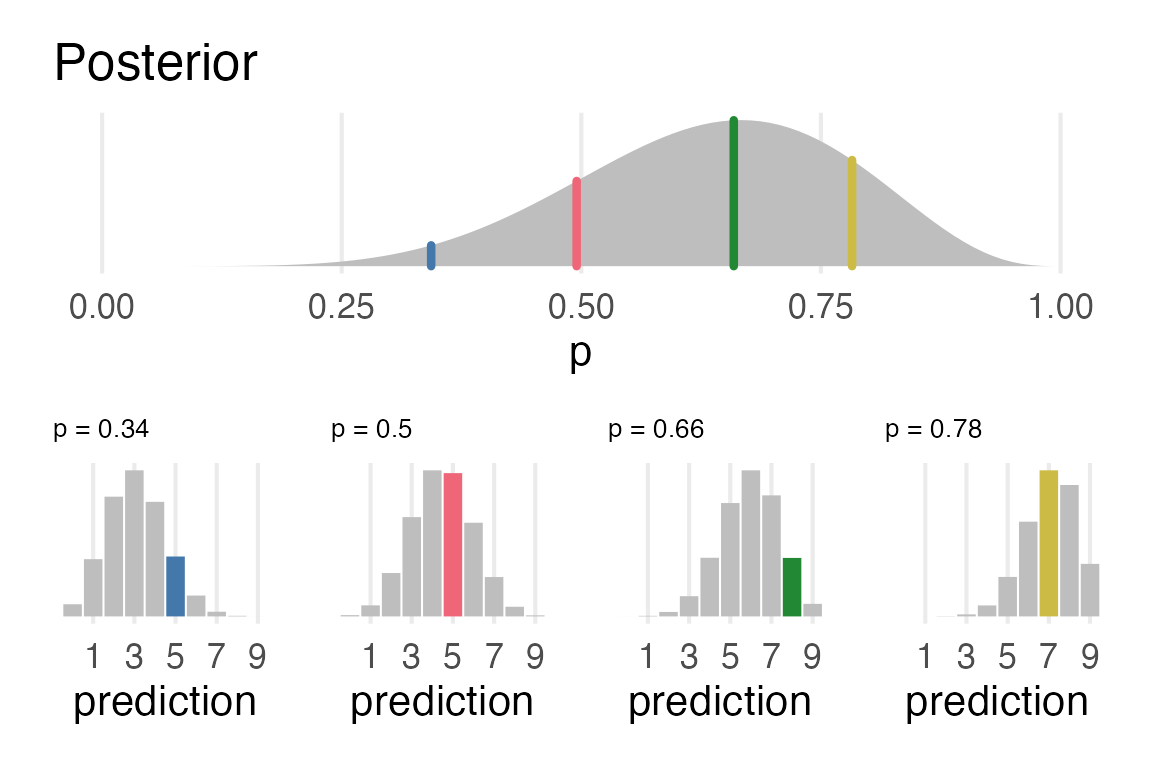
Pretty pleased with this!