I’m still thinking about priors, distributions, and logistic regressions. The fact that a fairly broad normal distribution in logit space turns into a bimodal distribution in probability space has got me thinking about the standard deviation of random effects in logistic regression. Specifically, what happens in cases where the population of individuals may be bimodal
Simulating some data
I’ll kick things off with simulating some data. Our predictor variable will be just randomly sampled from ~normal(0,1), so it’ll handily already be z-scored. I’ll also go for a slope in logit space of 1, so logit(y) = x.
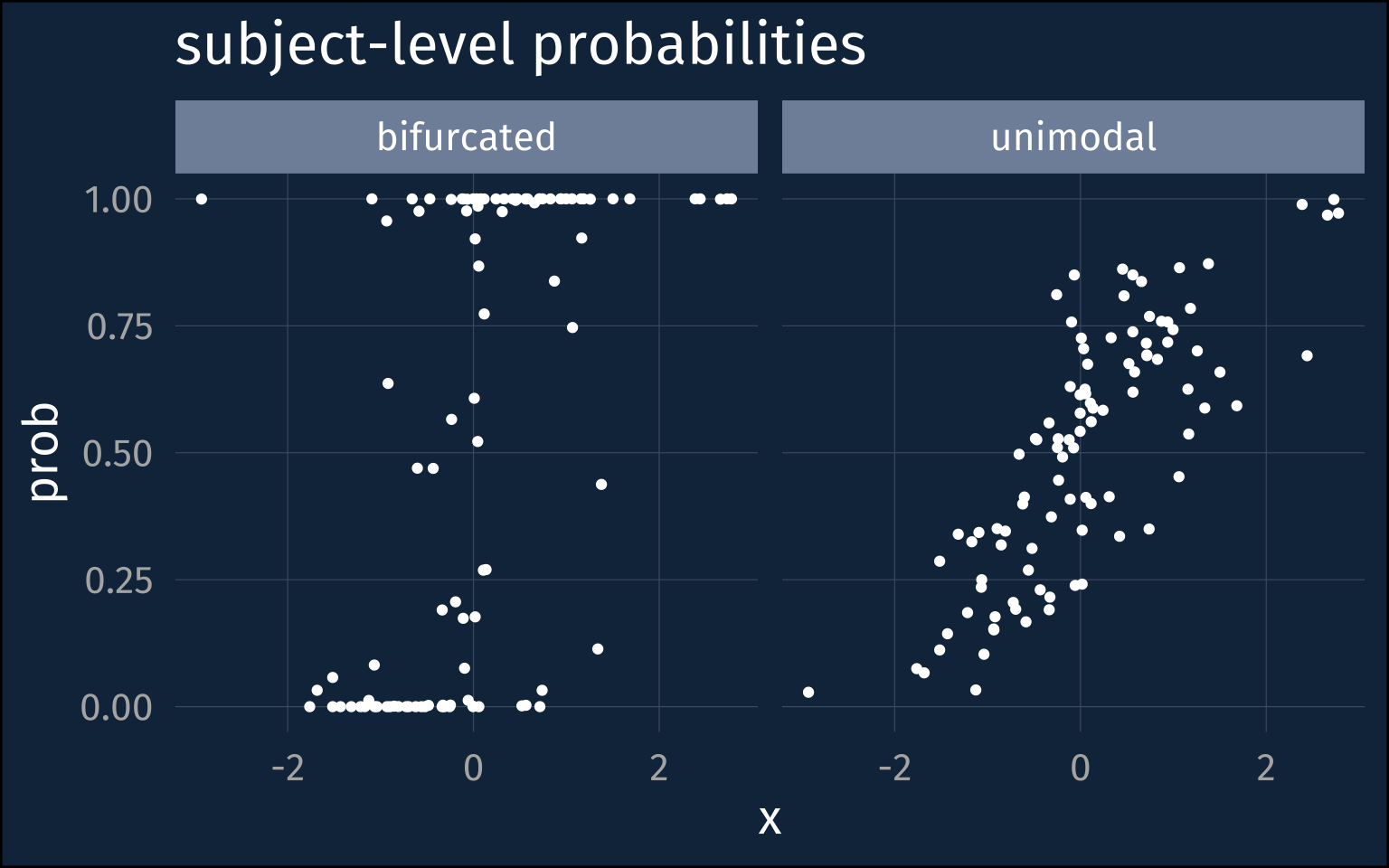
I’ll treat each point I’ve sampled for X as belonging to an individual, or subject, grouping variable, and each individual’s personal probability will be sampled from a beta distribution. I’ll simulate two possibilities here, one where individuals are kind of closely clustered near each other, and another where they’re pretty strongly bifurcated close to 0 and 1.1
plotting code
sim_params |>
pivot_longer(
starts_with("subj_prob")
) |>
mutate(
name = case_when(
name == "subj_prob_high" ~ "unimodal",
name == "subj_prob_low" ~ "bifurcated"
)
) |>
ggplot(aes(x, value))+
geom_point()+
labs(title = "subject-level probabilities",
y = "prob")+
scale_x_continuous(
breaks = seq(-2,2, by = 2)
) +
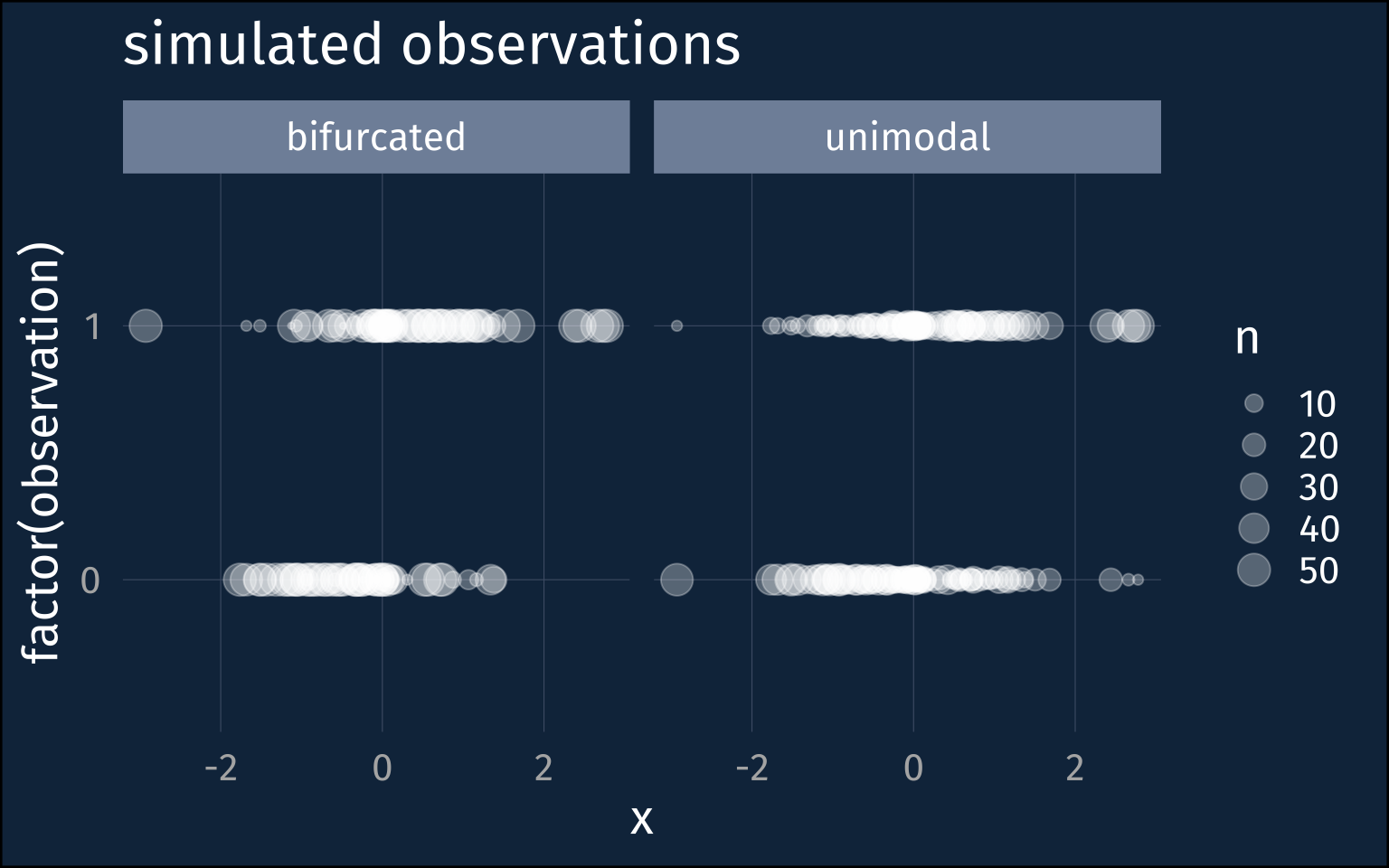
facet_wrap(~name)For each of these probabilities, I’ll simulate 50 binomial observations.
plotting code
sim_obs |>
pivot_longer(
starts_with("y_"),
names_to = "simulation",
values_to = "observation"
) |>
mutate(
simulation = case_when(
simulation == "y_prob_high" ~ "unimodal",
simulation == "y_prob_low" ~ "bifurcated"
)
) |>
ggplot(aes(x, factor(observation))) +
stat_sum(alpha = 0.3)+
labs(title = "simulated observations")+
scale_x_continuous(
breaks = seq(-2,2, by = 2)
) +
facet_wrap(~simulation)Looking at the default priors
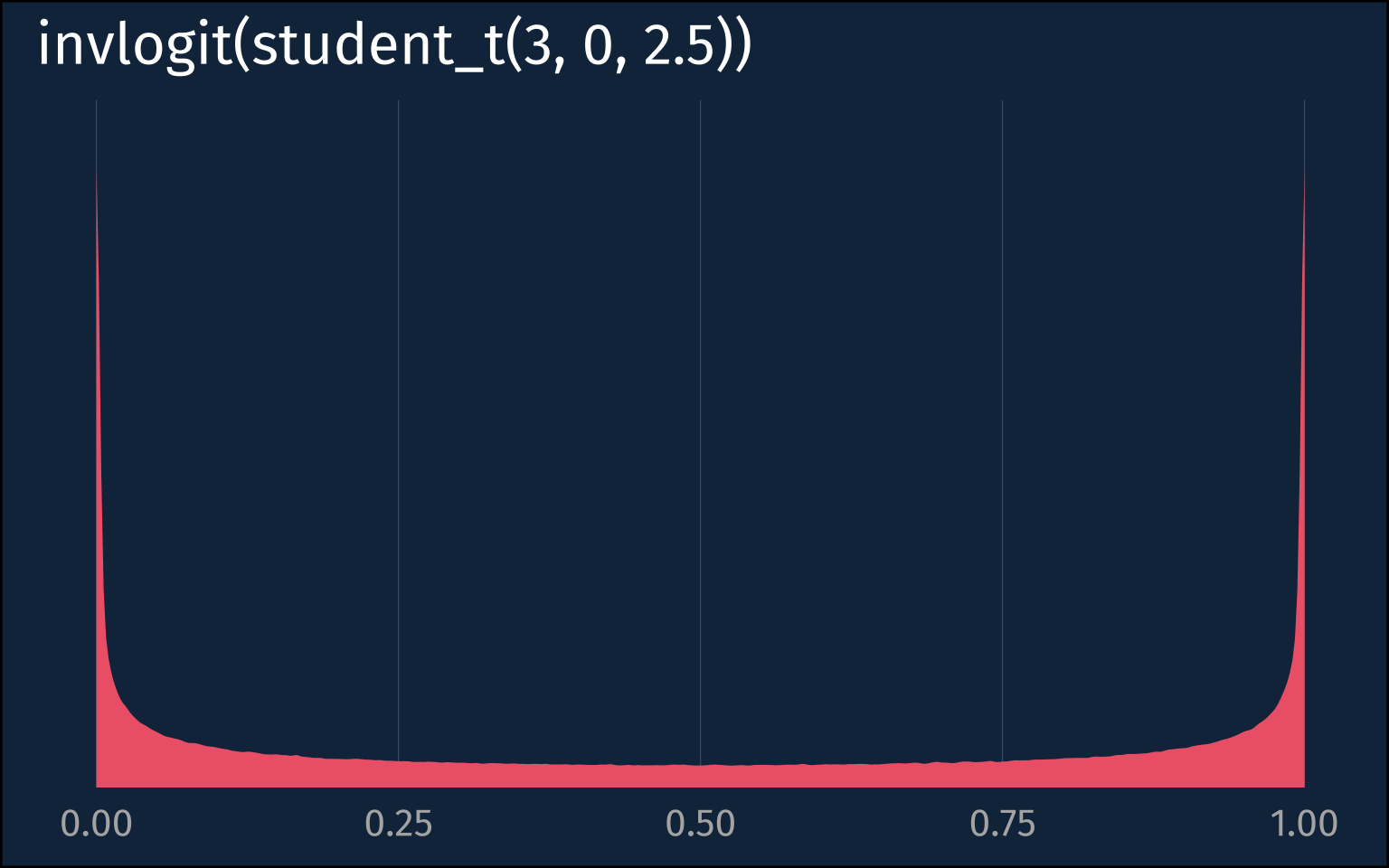
If we take a look at the default priors a logistic model would get in brms, we can see that both the Intercept and the slope get pretty broad priors (the blank prior for the slope means it’s a flat prior).
| prior | class | coef | group |
|---|---|---|---|
| b | |||
| b | x | ||
| student_t(3, 0, 2.5) | Intercept | ||
| student_t(3, 0, 2.5) | sd | ||
| sd | individual | ||
| sd | Intercept | individual |
If we real quick look at how the prior on the intercept plays out in the probability space, we get one of these bimodal distributions.
plotting code
tibble(
x = rstudent_t(1e6, df = 3, sigma = 2.5)
) |>
ggplot(aes(plogis(x))) +
stat_slab(fill = ptol_red)+
theme_no_y()+
scale_y_continuous(
expand = expansion(mult = 0)
)+
labs(title = "invlogit(student_t(3, 0, 2.5))",
x = NULL)So, for the intercept and slope priors, I’ll adjust them to be ~normal(0, 1.5) and ~normal(0,1), respectively.
Actually fitting the models.
Bimodal population
First, here’s the model for the population where individuals’ probabilities were squished out towards 0 and 1.
summary table
low_mod |>
gather_draws(
`sd_.*`,
`b_.*`,
regex = T
) |>
mean_hdci() |>
select(.variable, .value, .lower, .upper) |>
gt() |>
fmt_number()| .variable | .value | .lower | .upper |
|---|---|---|---|
| b_Intercept | 0.60 | −1.04 | 2.13 |
| b_x | 3.01 | 1.62 | 4.36 |
| sd_individual__Intercept | 7.85 | 5.85 | 10.21 |
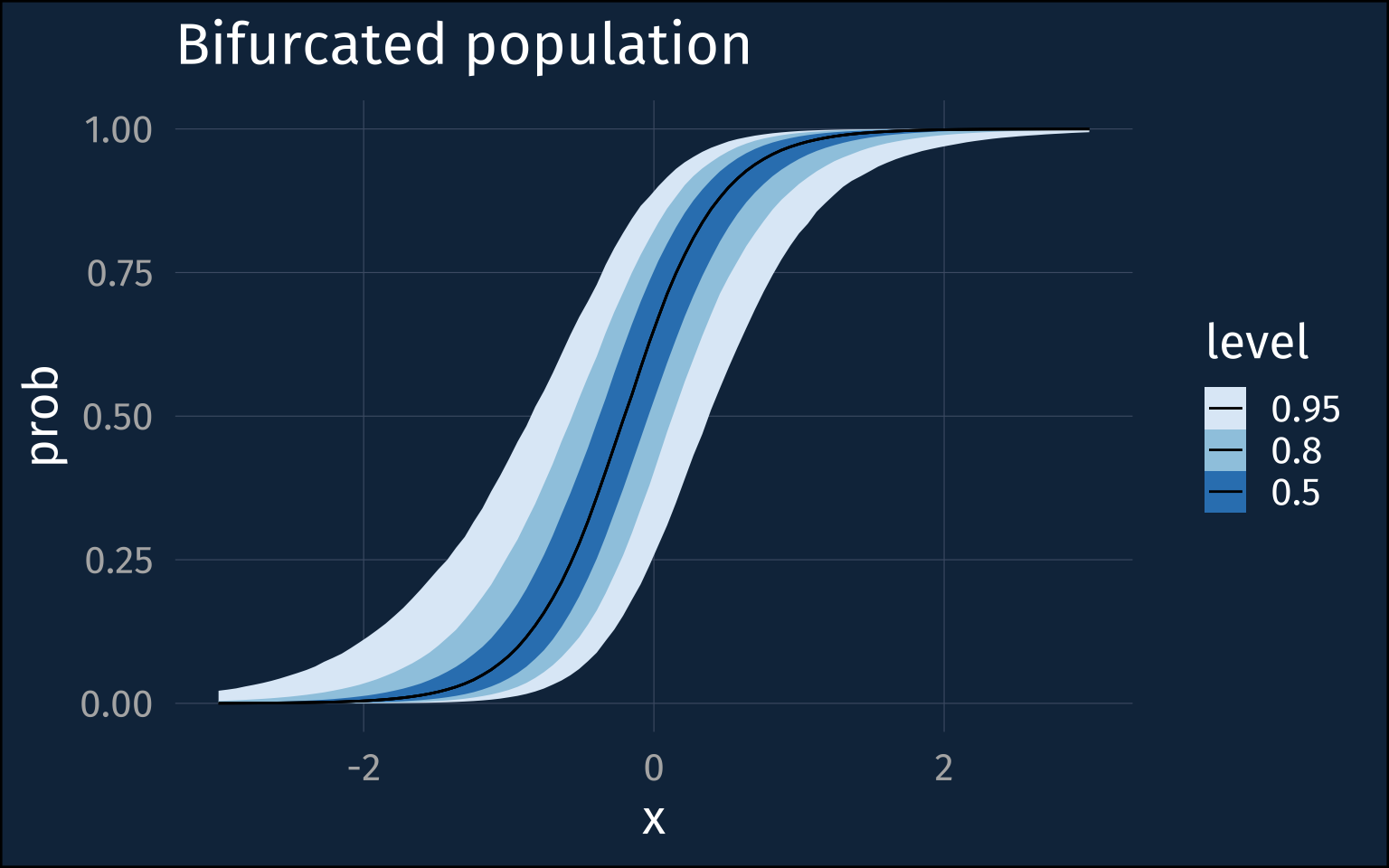
Things have kind of clearly gone off the rails here. The intercepts and slopes are all over the place, but that’s maybe not surprising given the trade offs the model is making between the population level slopes and the individual level probabilities. It’s worth noting that this model had no diagnostic warnings and was well converged.
plotting code
low_mod |>
predictions(
newdata = datagrid(x = seq(-3, 3, length = 100)),
re_formula = NA
) |>
posterior_draws() |>
ggplot(aes(x, draw)) +
stat_lineribbon(linewidth = 0.5)+
scale_fill_brewer() +
labs(
title = "Bifurcated population",
y = "prob"
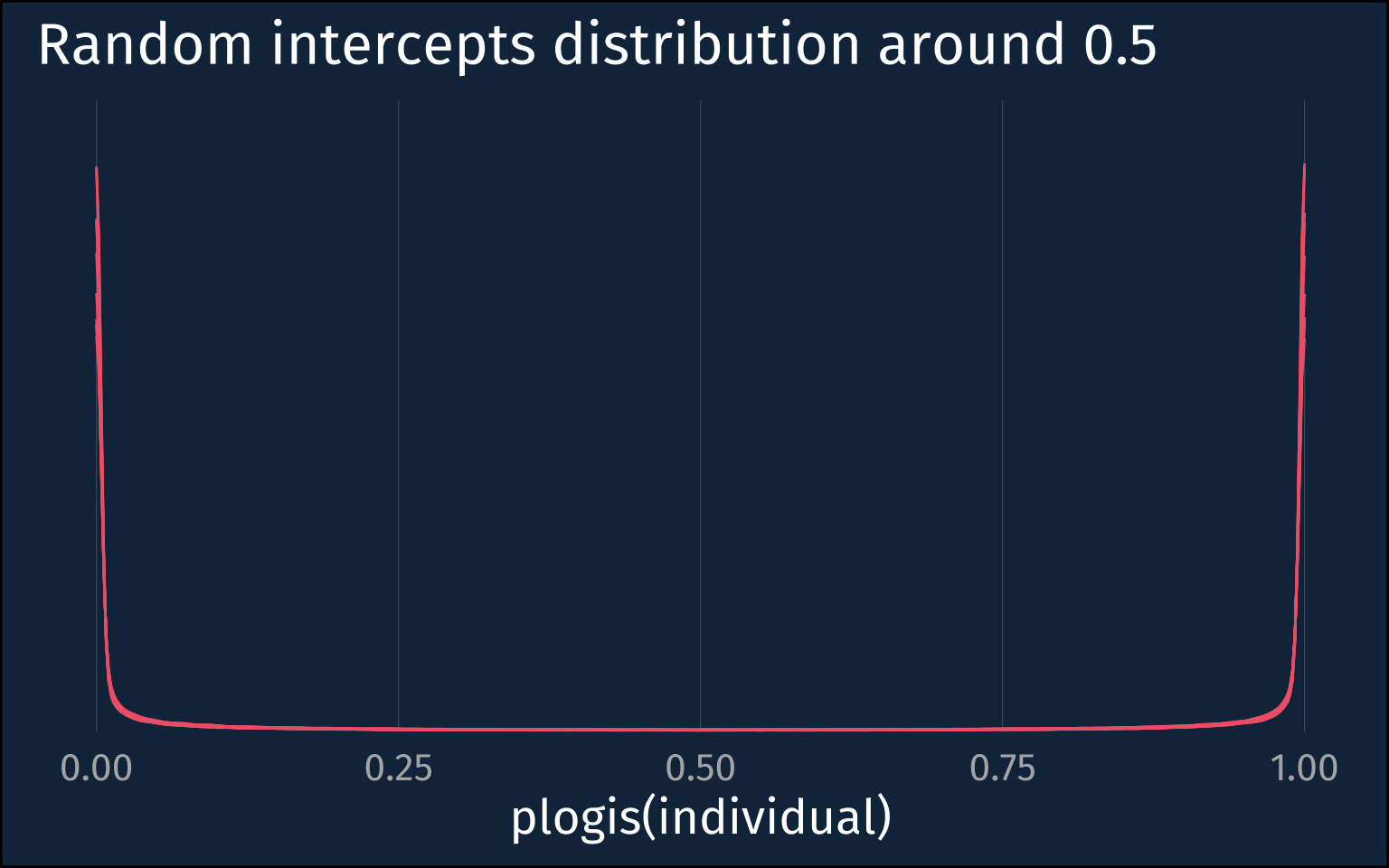
)In fact, that posterior distribution for the between-speaker sd is very extreme at about 8. If we plot the kind of distribution of individuals it suggests when the population level probability = 0.5, we get those steep walls near 0 and 1 again.
plotting code
low_mod |>
gather_draws(
`sd_.*`,
regex = T
) |>
slice_sample(
n = 10
) |>
rowwise() |>
mutate(
individuals = list(tibble(
individual = rnorm(1e5, mean = 0, sd=.value)
))
) |>
unnest(individuals) |>
ggplot(aes(plogis(individual)))+
stat_slab(
aes(group = factor(.value)),
linewidth = 0.5,
fill = NA,
color = ptol_red
) +
scale_y_continuous(expand = expansion(mult = 0))+
labs(
title = "Random intercepts distribution around 0.5"
)+
theme_no_y()The Unimodal Population
Let’s do it all again, but now for the population where individuals’ probabilities were clustered around the population probability.
summary table
high_mod |>
gather_draws(
`sd_.*`,
`b_.*`,
regex = T
) |>
mean_hdci() |>
select(
.variable, .value, .lower, .upper
) |>
gt() |>
fmt_number(decimals = 2)| .variable | .value | .lower | .upper |
|---|---|---|---|
| b_Intercept | −0.06 | −0.23 | 0.10 |
| b_x | 1.05 | 0.87 | 1.22 |
| sd_individual__Intercept | 0.77 | 0.63 | 0.91 |
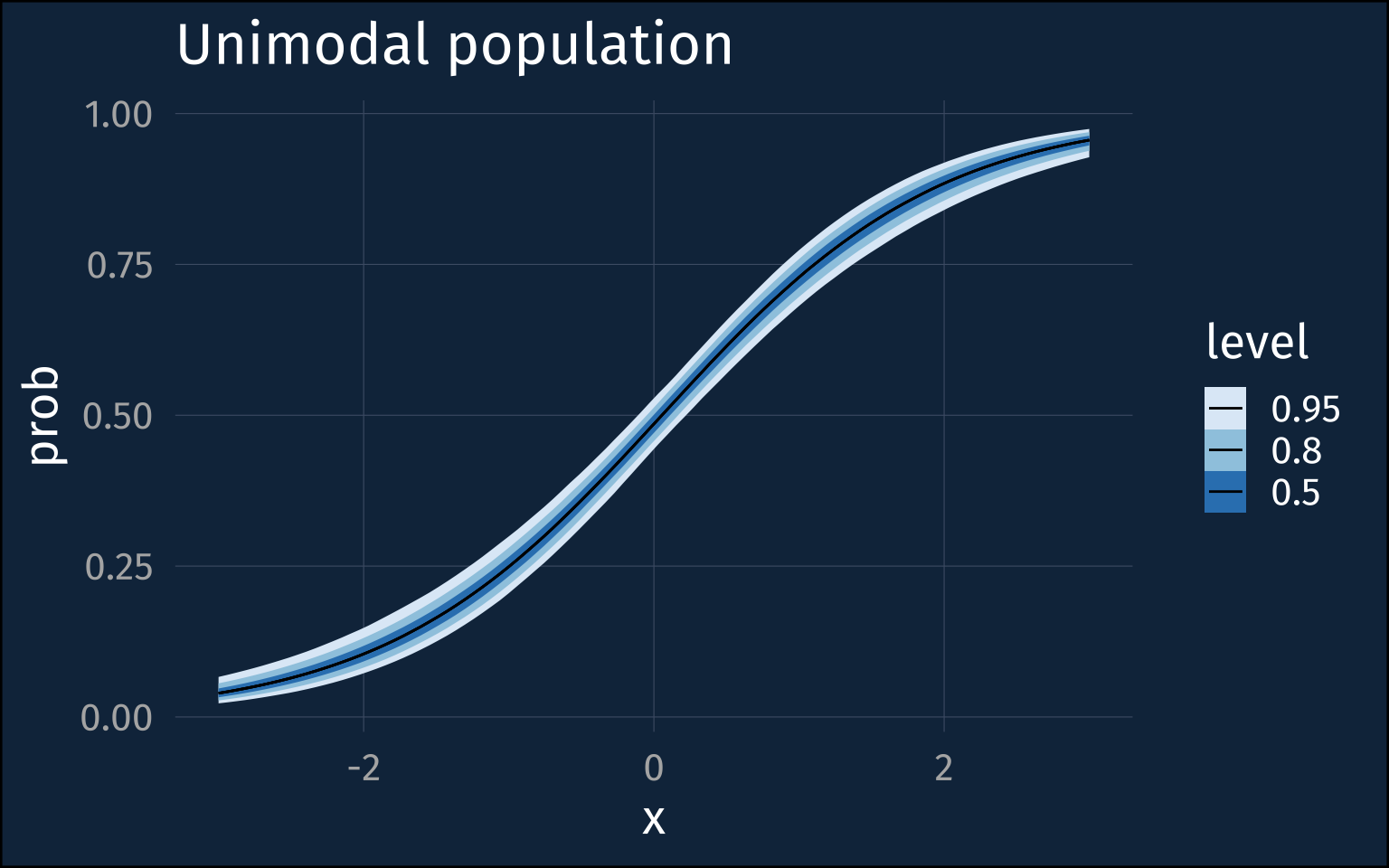
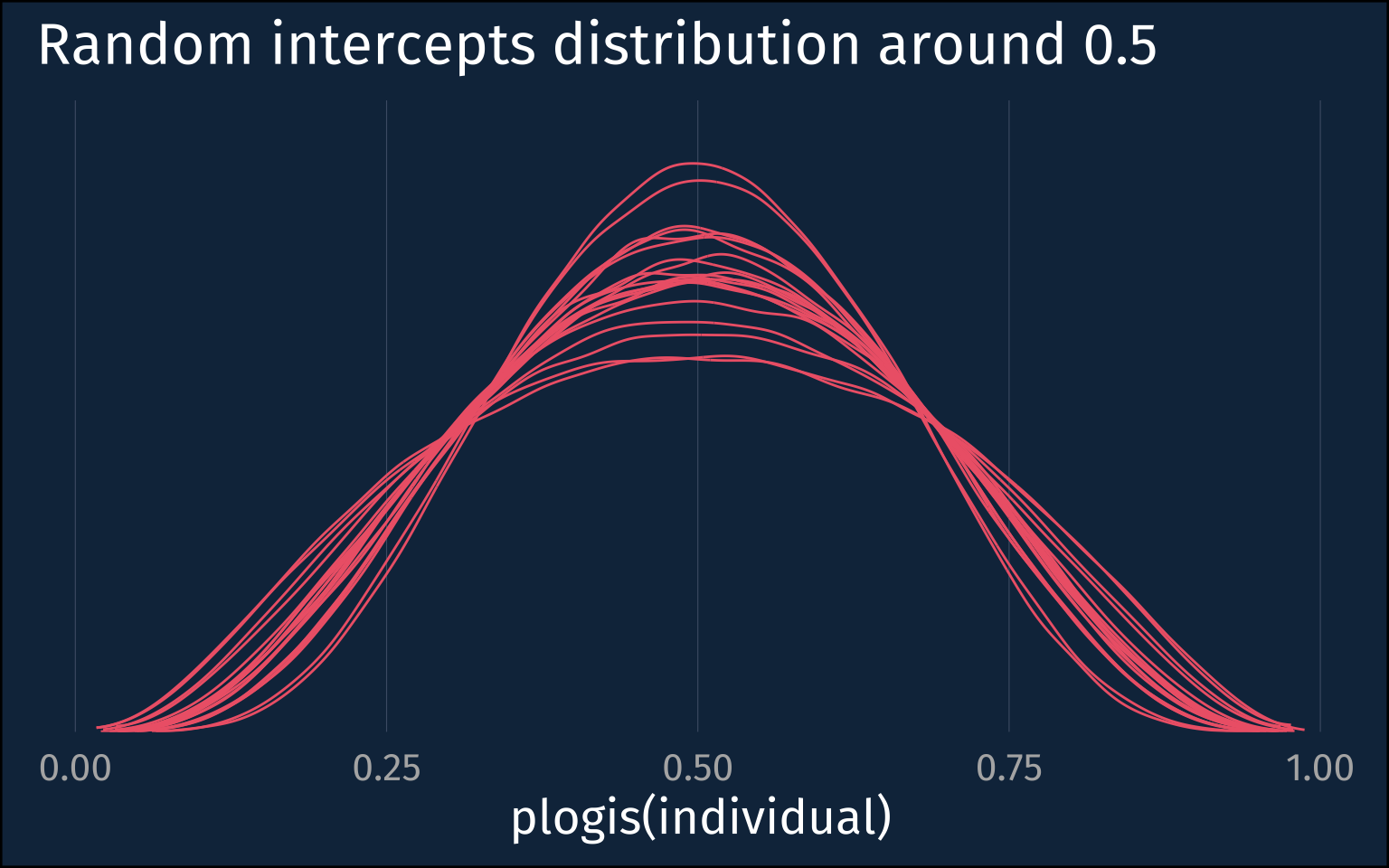
The intercepts and slope posteriors are much more tight, and the inter-speaker sd posterior is <1.
plottng code
high_mod |>
predictions(
newdata = datagrid(x = seq(-3, 3, length = 100)),
re_formula = NA
) |>
posterior_draws() |>
ggplot(aes(x, draw)) +
stat_lineribbon(linewidth = 0.5)+
scale_fill_brewer()+
labs(
title = "Unimodal population",
y = "prob"
)Let’s look at the implied individual level distribution around 0.5.
plotting code
high_mod |>
gather_draws(
`sd_.*`,
regex = T
) |>
slice_sample(
n = 20
) |>
rowwise() |>
mutate(
individuals = list(tibble(
individual = rnorm(1e5, mean = 0, sd=.value)
))
) |>
unnest(individuals) |>
ggplot(aes(plogis(individual)))+
stat_slab(
aes(group = factor(.value)),
linewidth = 0.5,
fill = NA,
color = ptol_red
) +
scale_y_continuous(expand = expansion(mult = 0))+
labs(
title = "Random intercepts distribution around 0.5"
)+
theme_no_y()The Upshot
Even though I’ve fit and looked at hierarchical logistic regressions before, I hadn’t stopped to think about how to interpret the standard deviation of the random intercepts before. If you’d asked me before what a large sd implied about the distribution of individuals in the probability space, I think I would have said they’d be more uniformly distributed, but actually it means they’re more bifurcated!
Also, if you’ve got a fairly bifurcated population, the population level estimates are going to get pretty wonky.
All food for thought moving forward.
Footnotes
I initially also tried simulating a case where individuals were strictly categorical based on the probability associated with their x, and things did not go well for the models.↩︎
Reuse
Citation
@online{fruehwald2023,
author = {Fruehwald, Josef},
title = {Thinking {About} {Hierarchical} {Variance} {Parameters}},
series = {Væl Space},
date = {2023-06-29},
url = {https://jofrhwld.github.io/blog/posts/2023/06/2023-06-29_hierarchical-variance/},
doi = {10.59350/9gmbj-4f042},
langid = {en}
}