word_rt <- read_csv("https://raw.githubusercontent.com/bodowinter/applied_statistics_book_data/master/ELP_length_frequency.csv")Rows: 33075 Columns: 4
── Column specification ────────────────────────────────────────────────────────
Delimiter: ","
chr (1): Word
dbl (3): Log10Freq, length, RT
ℹ Use `spec()` to retrieve the full column specification for this data.
ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.Plots
Making plots of the data before we start modelling:
word_rt |>
ggplot(aes(Log10Freq, RT)) +
#geom_point()
stat_bin_2d()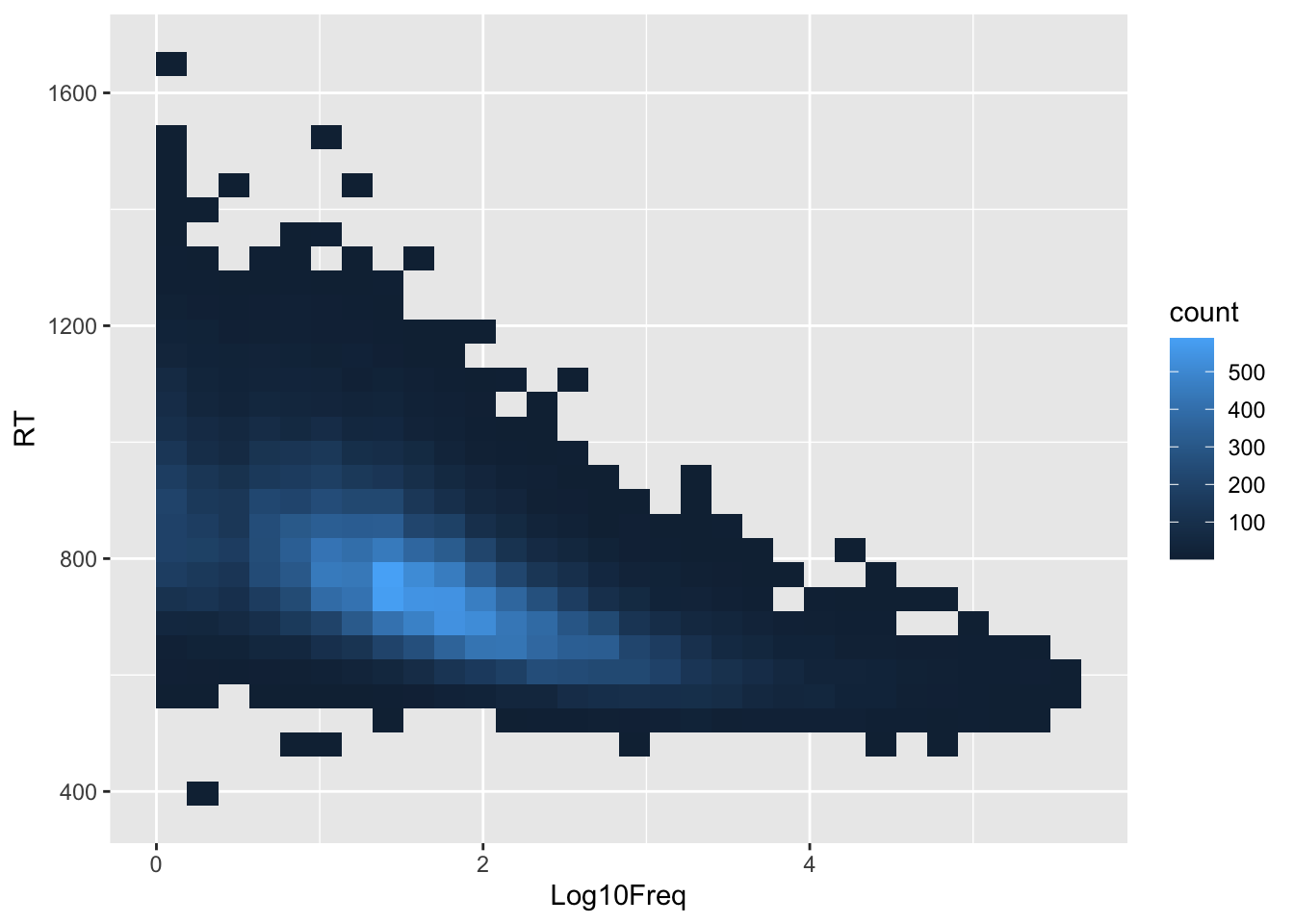
Looks like a negative effect of word frequency on reaction time
word_rt |>
ggplot(aes(length, RT)) +
stat_bin_2d()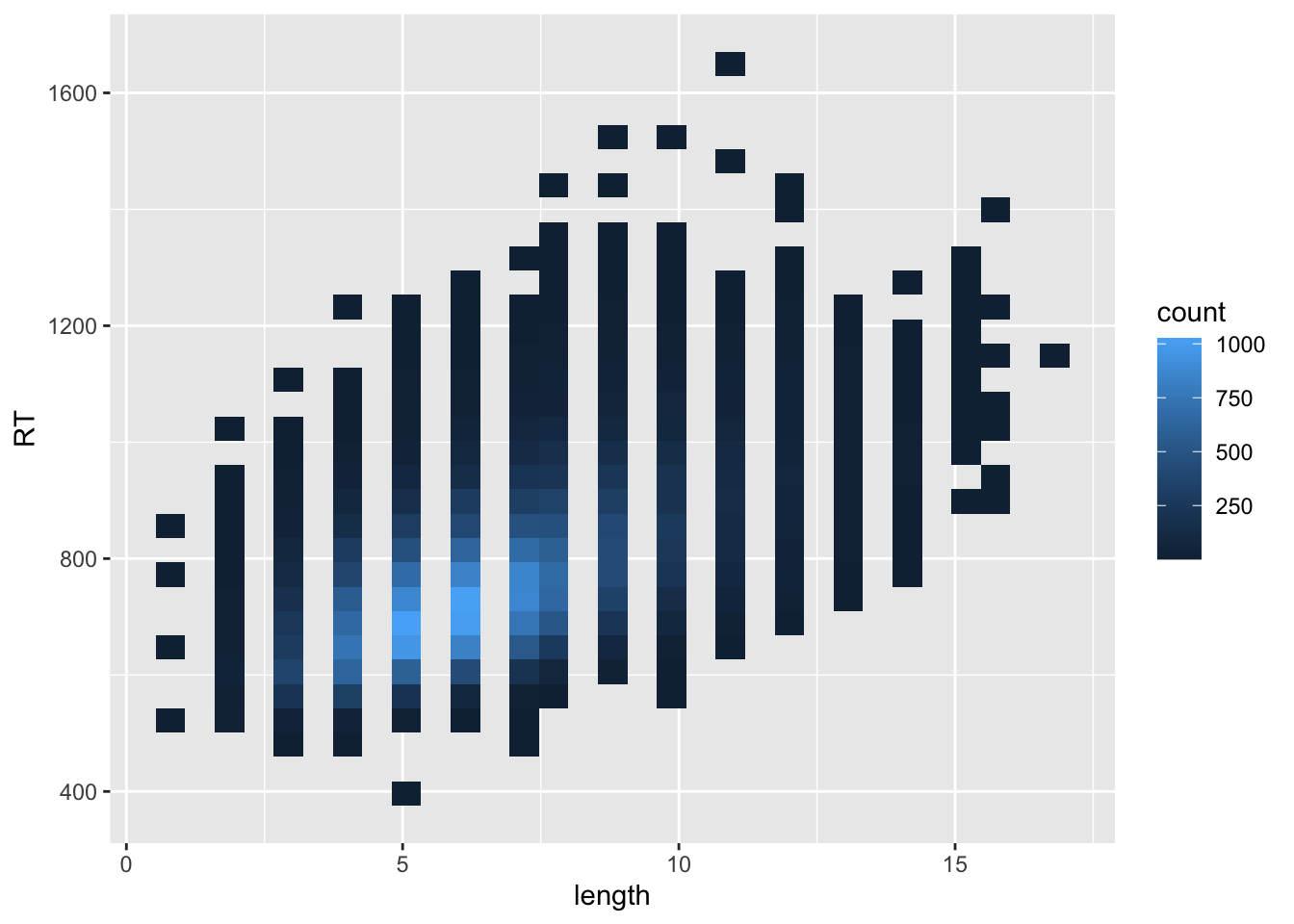
Looks like a positive effect of word length on reaction time.
word_rt |>
ggplot(aes(Log10Freq, length)) +
stat_bin_2d()+
stat_smooth()`geom_smooth()` using method = 'gam' and formula = 'y ~ s(x, bs = "cs")'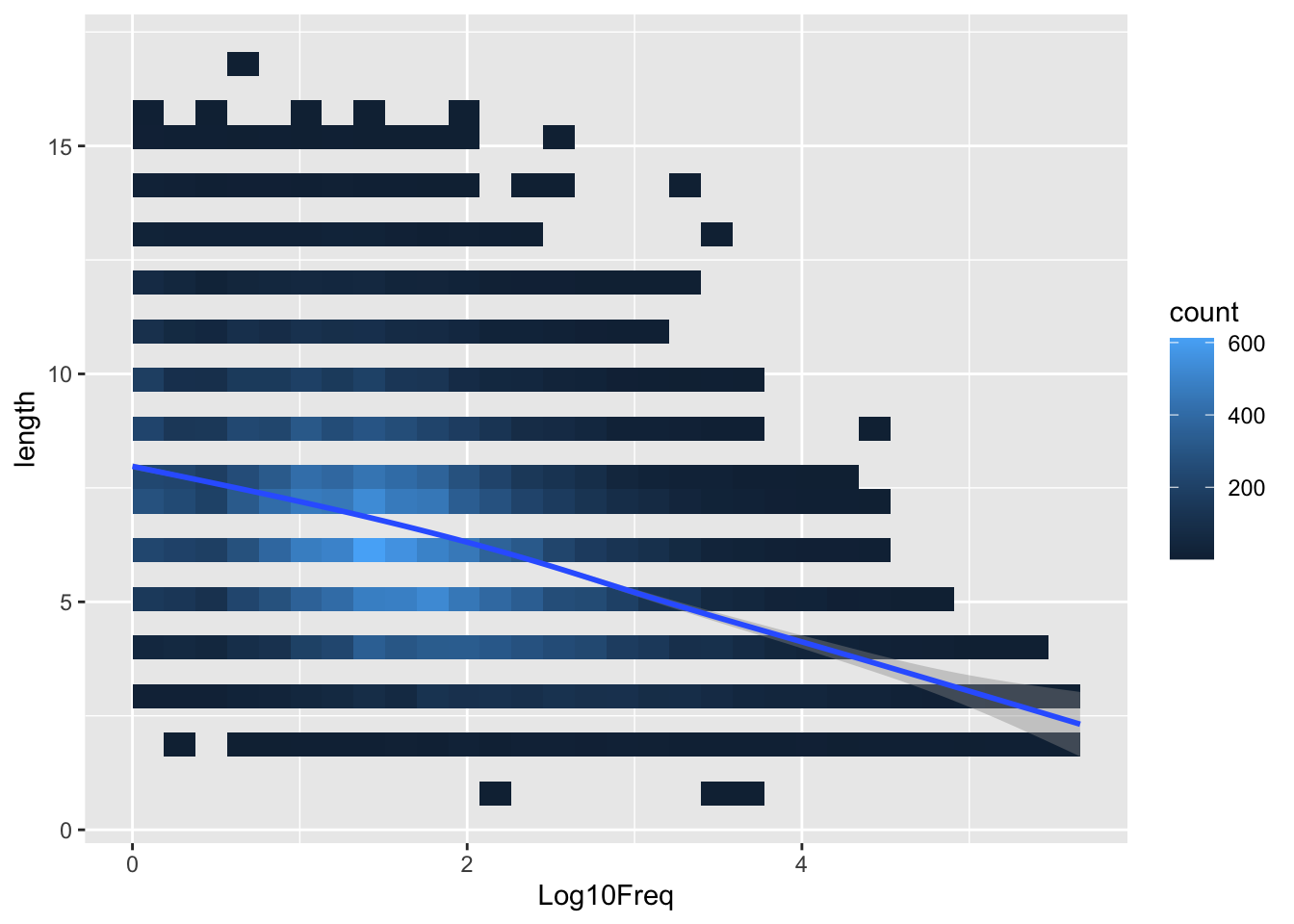
Looks like word frequency and word length are slightly collinear.
Not ideal (modelling without centering and scaling)
lm fits the model.
model_0 <- lm(RT ~ Log10Freq + length, data = word_rt)tidy(model_0)# A tibble: 3 × 5
term estimate std.error statistic p.value
<chr> <dbl> <dbl> <dbl> <dbl>
1 (Intercept) 748. 2.18 344. 0
2 Log10Freq -68.0 0.594 -115. 0
3 length 19.5 0.238 81.9 0The (Intercept) value is the predicted value of RT when both Log10Freq and length are 0. Not a likely combination of values.
Goodness of fit:
glance(model_0)# A tibble: 1 × 12
r.squared adj.r.sq…¹ sigma stati…² p.value df logLik AIC BIC devia…³
<dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 0.487 0.487 89.1 15717. 0 2 -1.95e5 3.91e5 3.91e5 2.62e8
# … with 2 more variables: df.residual <int>, nobs <int>, and abbreviated
# variable names ¹adj.r.squared, ²statistic, ³devianceScaling and Centering
We’ll center both predictors on their means, and scale by the standard deviation:
Let’s actually fit three models,
- One for just frequency
- One for just length
- One for frequency and length
We can examine all of these models goodness of fits together.
list(
freq = model_f,
length = model_l,
freq_length = model_fl
) |>
map_dfr(glance, .id = "model") |>
select(model, r.squared, adj.r.squared, AIC, BIC)# A tibble: 3 × 5
model r.squared adj.r.squared AIC BIC
<chr> <dbl> <dbl> <dbl> <dbl>
1 freq 0.383 0.383 396956. 396981.
2 length 0.284 0.284 401905. 401930.
3 freq_length 0.487 0.487 390856. 390889.The first thing to notice is that even though we’ve centered and scaled the data, the goodness of fit metrics for the freq_length mode, (r-squared and adjusted r-squared) are identical to the original multivariate model.
Also, the goodness of fit always gets better when we add a new predictor, but the adjusted r squared, AIC and BIC try to balance out the complexity of the model and the goodness of fit.
Evaluating the model
We can merge information from the model onto the original data with augment().
augment(model_fl, word_rt_scaled) |>
ggplot(aes(.fitted, .resid))+
geom_point()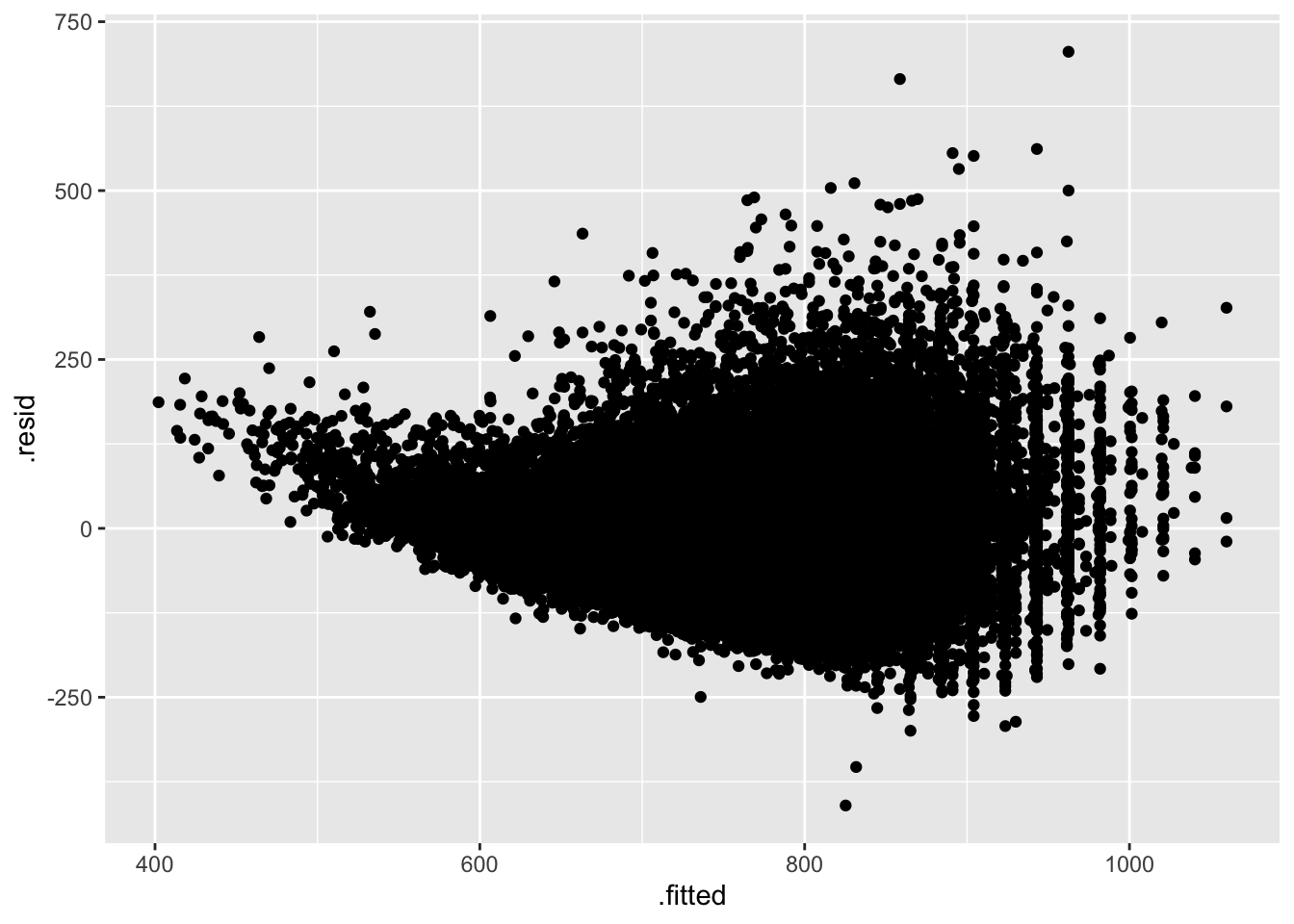
We can also look at the predicted values with marginaleffects::predictions()
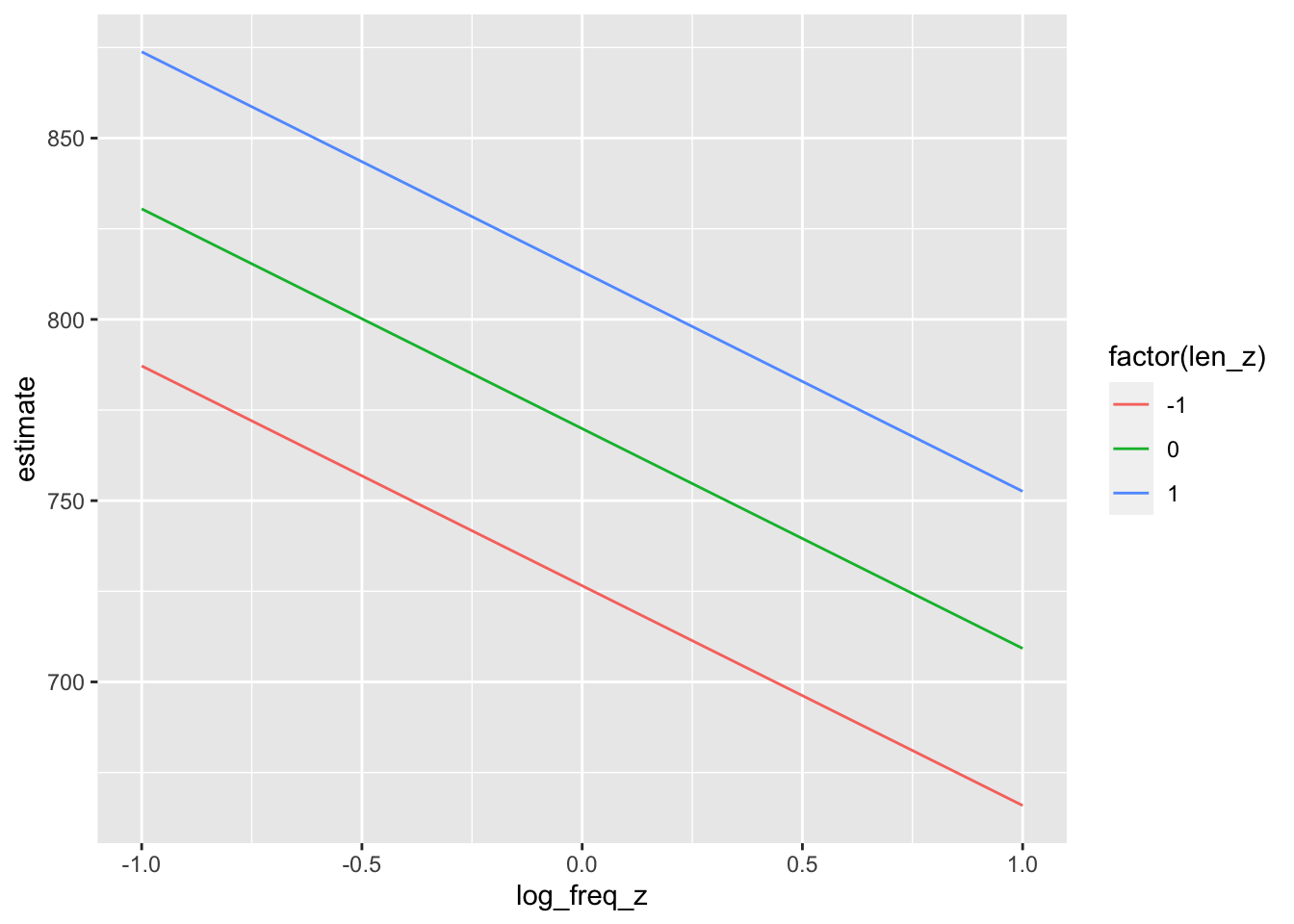
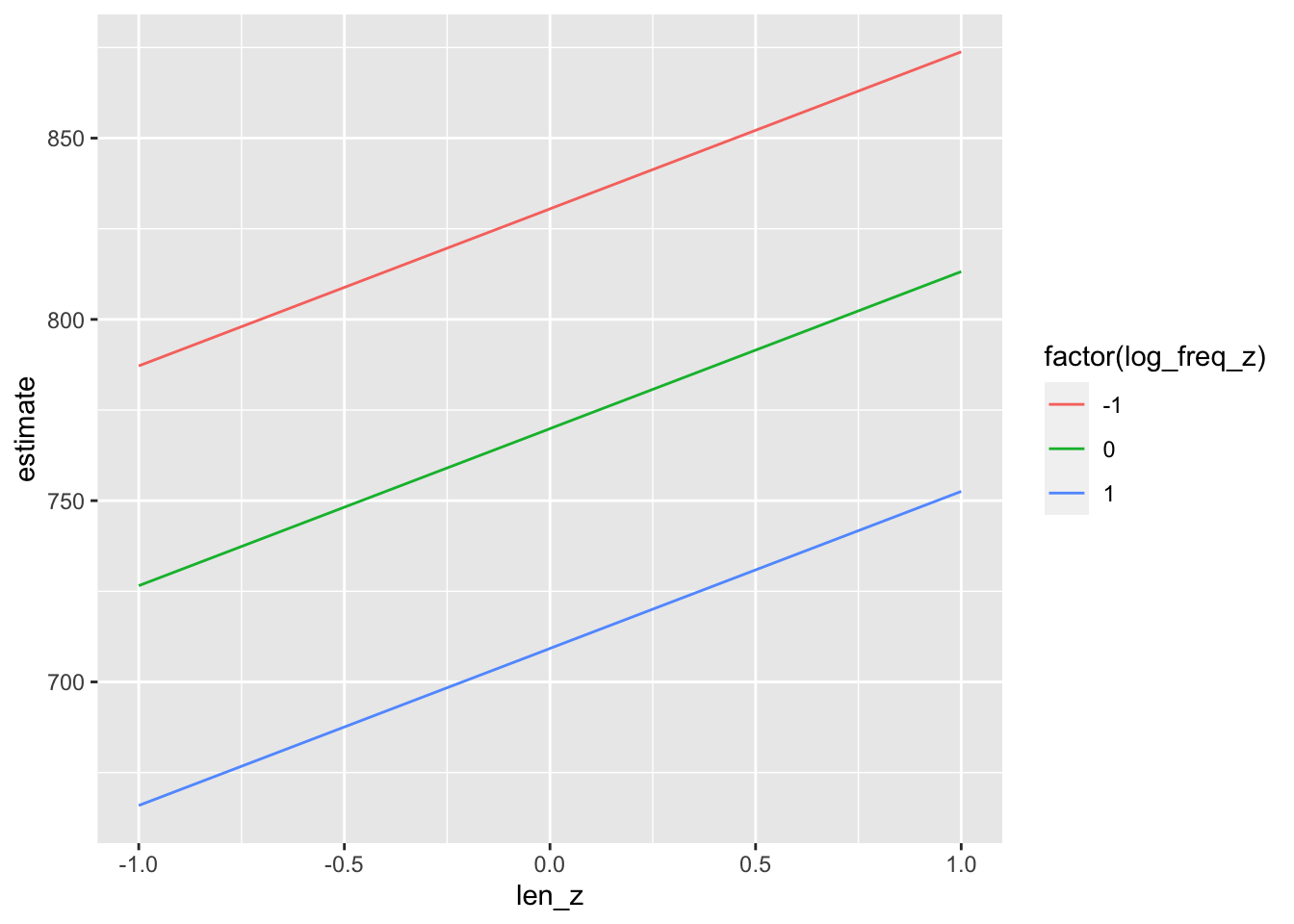
predictions(model_fl,
newdata = datagrid(log_freq_z = c(-1, 0, 1),
len_z = c(-1, 0, 1))) |>
tibble()->
predictions_fl